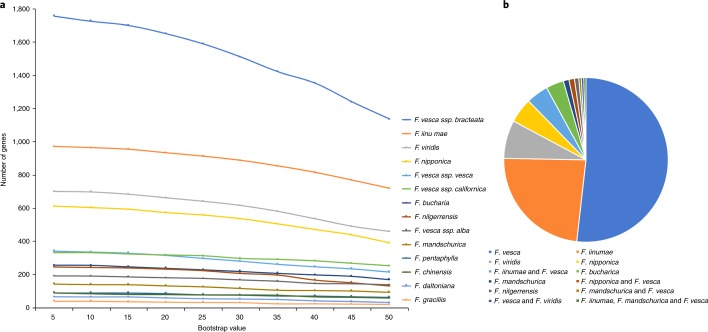
Fig. 1. Phylogenetic analyses.
a, Number of genes from species identified as being sister to a homoeolog from the octoploid genome, by using PhyDS with bootstrap support value (BSV) cutoffs. Based on previous results4. b, Reanalysis of the data, including in-paralogs and BSV50 cutoff, identified the same progenitor species. The prevalence and biased patterns of homoeologous exchanges between subgenomes resulted in the dominant F. vesca subgenome replacing a greater number of corresponding regions in each of the recessive subgenomes4. Thus, a greater number of genes from the dominant F. vesca subgenome were identified, with the F. iinumae–like subgenome being second.