Figure 4.
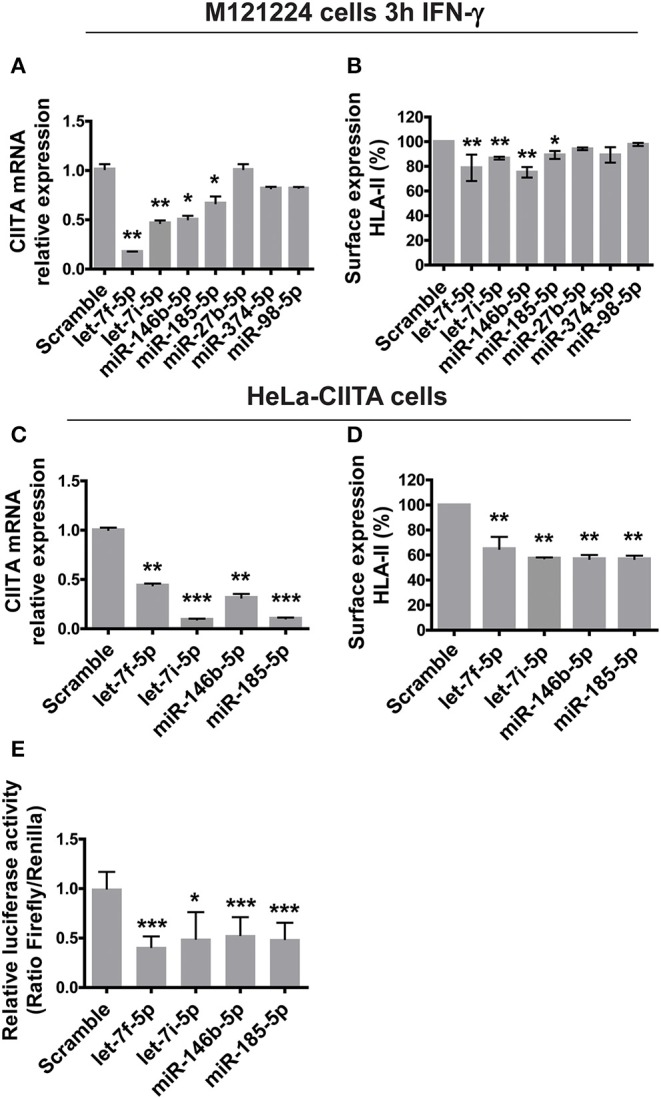
Functional analysis of miRNAs modulated by H. pylori. (A) Relative expression of mRNA encoding CIITA in M121224 melanoma cells activated for 3 h with 50 ng/ml IFN-γ and then transfected with pCMV-miR plasmid carrying the indicated miRNAs for 48 h. Data were normalized to an endogenous reference gene (β-actin). Values of cells expressing miRNA scramble were taken as reference and set as 1. (B) Surface expression of HLA-II in M121224 cells treated as in (A). HLA-II expression in transfected cells was relative to control cells (expressing miRNA scramble). Data are expressed as mean ± SEM of three independent experiments, performed with 3 different cell preparations. (C) Relative expression of mRNA encoding CIITA in HeLa-CIITA cells transfected with pCMV-miR plasmid carrying the indicated miRNAs for 48 h. Data were normalized to an endogenous reference gene (β-actin). Values of cells expressing miRNA scramble were taken as reference and set as 1. (D) Surface expression of HLA-II in HeLa-CIITA cells treated as in (C). HLA-II expression in transfected cells was relative to control cells. Data are expressed as mean ± SEM of three independent experiments, performed with three different cell preparations. (E) Luciferase assay. Luciferase assay supports the interaction between let-7f-5p,−7i-5p, miR-146b-5p, and -185-5p and the 3′-UTR of CIITA. As controls, we used cells transfected with pCMV-miR plasmid encoding miRNA scramble. Data are expressed as mean ± SEM of three independent experiments. Significance was determined by Student's t-test. *p < 0.05; **p < 0.01; and ***p < 0.001.
