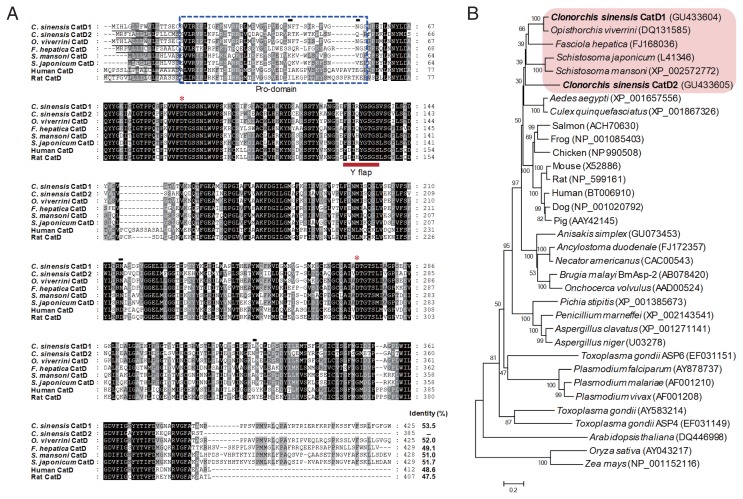
Fig. 1.
Multiple sequence alignment and phylogenetic analyses. (A) Multiple sequence analysis. The deduced amino acid sequences of 2 CsCatDs were aligned with those of Opisthorchis viverrini (AAZ39883), Fasciola hepatica (ABJ97284), Schistosoma mansoni (AAB63442), Schistosoma japonicum (AAC37302), Human (NP_001900), and Rat (NP_599161). Gaps are introduced into the sequences to maximize alignment. The asterisks indicate the catalytic aspartic acid residues. Black lines under the sequences are predicted signal peptide sequences. Potential pro-peptide regions are marked with a blue dotted box. The Y flap element is indicated by a bold red line. Predicted N-glycosylation sites in CsCatDs are represented by black lines. The shading represents the degree of identity among the sequences: black (100%), dark grey (50–92%), and light grey (15–49%). (B) Phylogenetic analysis. The phylogenetic tree was constructed with the neighbor-joining method using the MEGA4 program. Numbers on the branches indicate bootstrap proportions (1,000 replicates).