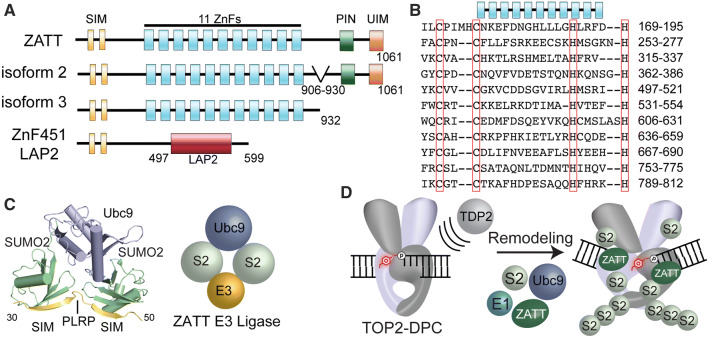
Fig. 5.
ZATT domain architecture and SUMOylation. a Four splicing isoforms ZnF451: ZATT (isoform1), isoform2 and 3 and ZnF451 Lap2a. ZATT domain structure consists of 2 N-terminal SUMO interacting motifs (SIMs), 11 Zinc Finger domains (ZnFs), PilT N-terminal domain (PIN) NYN domain, and a Ubiquitin Interacting Motif (UIM). b Uniprot alignment of predicted central ZnF domains of ZATT. c X-ray Crystal structure PDB ID: 5D2 M. ZNF451 in complex with SUMOylation machinery, SUMO2 and the E2 Ligase, Ubc9. Znf451 dual SIM domain and PLRP is composed of amino acids 30-50 bound to SUMO2. d Model of ZATT interaction in TOP2-DPC resolution. ZATT, an E3 ligase, along with other SUMOylation machinery (E1 ligase, E2 ligase (human Ubc9/yeast Sae2) and SUMO2 (S2) allows for SUMOylation of TOP2-DPCs. ZATT binds to TOP2-DPC allowing for remodeling and TDP2 accessibility to the 5′Y