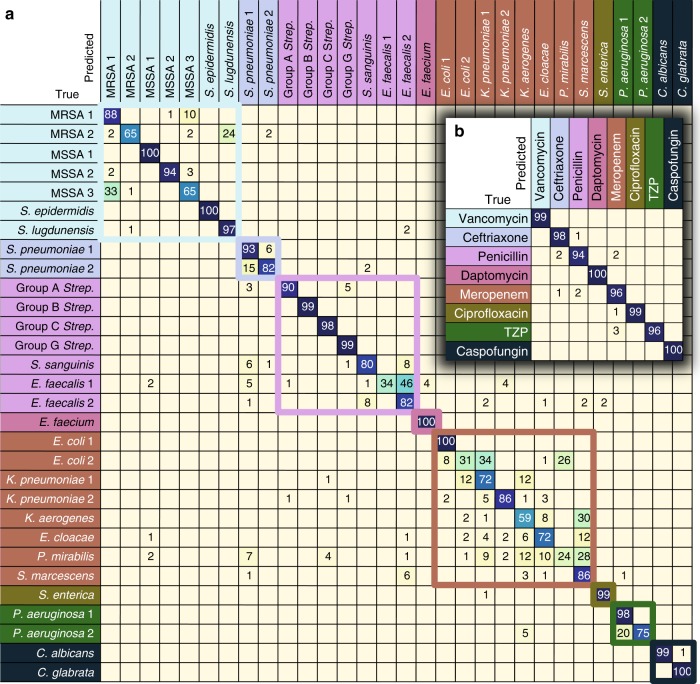
Fig. 2.
CNN performance breakdown by class. The trained CNN classifies 30 bacterial and yeast isolates with isolate-level accuracy of 82.2±0.3% and antibiotic grouping-level accuracy of 97.0±0.3% (± calculated as standard deviation across 5 train and validation splits). a Confusion matrix for 30 strain classes. Entry i, j represents the percentage out of 100 test spectra that are predicted by the CNN as class j given a ground truth of class i; entries along the diagonal represent the accuracies for each class. Misclassifications are mostly within antibiotic groupings, indicated by colored boxes, and thus do not affect the treatment outcome. Values below 0.5% are not shown, and matrix entries covered by figure insets are all below 0.5% aside from a 2% misclassification of MRSA 2 as P. aeruginosa 1 and 1% misclassification of Group B Strep. as K. aerogenes. b Predictions can be combined into antibiotic groupings to estimate treatment accuracy. TZP = piperacillin-tazobactam. All values below 0.5% are not shown