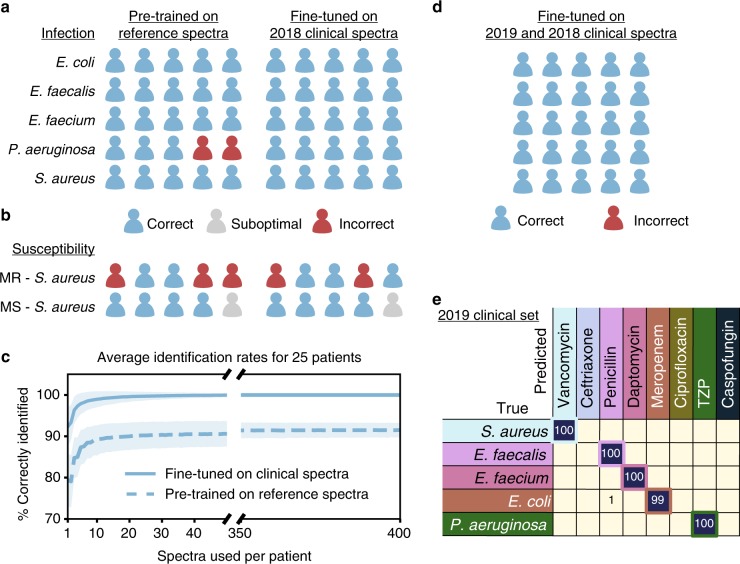
Fig. 4.
Extension to clinical patient isolates. A CNN pre-trained on our reference dataset can be extended to classify clinical patient isolates and further improved by fine-tuning on a small number of clinical spectra. a 5 species of bacterial infections are tested, with 5 patients per infection type. Each patient is classified into one of 8 treatment classes where each species corresponds to a different treatment class. After fine-tuning, species identification accuracy improves from 89.0±3.6% to 99.0±1.9% (± calculated as standard deviation across 10,000 sampling trials). b Binary classification between MRSA and MSSA patient isolates is also performed, with an accuracy of 61.7±7.3% that improves to 65.4±6.3% after fine-tuning. c Dependence of average diagnosis rates for the fine-tuned model on the number of spectra used per patient. With just 10 spectra, the performance of the model reaches 99% — within 1% difference of the performance with 400 spectra (100%). Error bars are calculated as the standard deviation across 10,000 trials of random selections of n spectra, where n is the number of spectra used per patient. d We perform an additional test on a new clinical dataset gathered from an additional 25 patients with the same distribution across species as the first clinical dataset. We update the model that is pre-trained on the reference dataset and fine-tuned on the first clinical dataset by fine-tuning on the second clinical dataset using the same procedure. e Detailed breakdown by class for the second clinical dataset. Correct pairings between species and treatment group are outlined in the colored boxes. The rate of accurate identification is 99.7±1.1%