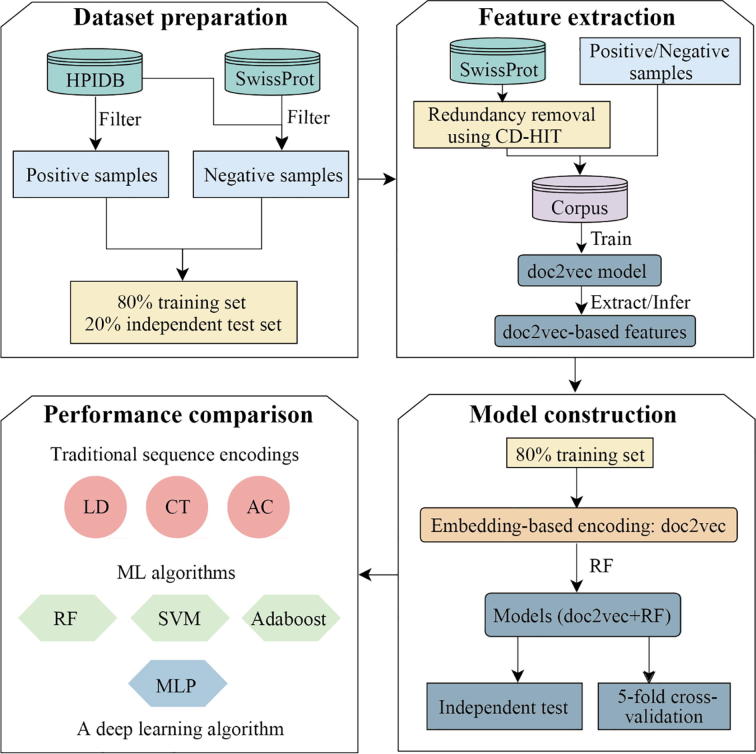
Fig. 1.
Workflow of our computational pipeline to predict human-virus PPIs. In the dataset preparation step, we constructed positive and negative data samples, utilizing human-virus protein interaction data from HPIDB as well as SwissProt database. Furthermore, we randomly sampled 80% as training data, while remining data was used as an independent test set. In the feature extraction step, we formed a corpus of sequence information from such protein data to train a doc2vec model, allowing us to extract/infer protein sequence specific features. Representing 80% of interactions between proteins through such feature embeddings as training data we used Random Forests (RF) to predict protein interactions using 5-fold cross-validation and independent test sets (remaining 20% of interaction data). In the final step, we compared our doc2vec + RF model with combinations of different encoding schemes such as the Conjoint Triad (CT), Local Descriptor (LD) and Auto Covariance (AC) and widely used ML methods such as Support Vector Machine (SVM), Multiple Layer Perceptron (MLP) and Adaptive Boosting (Adaboost).