Abstract
In light microscopy, illuminating light is passed through the sample as uniformly as possible over the field of view. For thicker samples, where the objective lens does not have sufficient depth of focus, light from sample planes above and below the focal plane will also be detected. The out-of-focus light will add blur to the image reducing the resolution. In fluorescence microscopy, any dye molecules in the field of view will be stimulated, including those in out-of-focus planes. Confocal microscopy provides a means of rejecting the out-of-focus light from the detector such that it does not contribute blur to the images being collected. This technique allows for high-resolution imaging in thick tissues.
In a confocal microscope, the illumination and detection optics are focused on the same diffraction-limited spot in the sample, which is the only spot imaged by the detector during a confocal scan. To generate a complete image, the spot must be moved over the sample and data collected point by point. A significant advantage of the confocal microscope is the optical sectioning provided, which allows for 3D reconstruction of a sample from high-resolution stacks of images. Several types of confocal microscopes have been developed for this purpose and each has different advantages and disadvantages. This paper provides a concise introduction to confocal microscopy.
Keywords: confocal microscopy, fluorescence, laser scanning, resonant scanning, spinning disk
Overview
The primary functions of a confocal microscope are to produce a point source of light and reject out-of-focus light, which provides the ability to image deep into tissues with high resolution, and optical sectioning for 3D reconstructions of imaged samples. The basic principle of confocal microscopy is that the illumination and detection optics are focused on the same diffraction-limited spot, which is moved over the sample to build the complete image on the detector. While the entire field of view is illuminated during confocal imaging, anything outside the focal plane contributes little to the image, lessening the haze observed in standard light microscopy with thick and highly-scattering samples, and providing optical sectioning.
The idea of rejecting out-of-focus light in this manner was patented in the 1950s by Marvin Minsky (Minsky, 1957; 1988) and achieved by the use of illumination - and detection-side pinhole apertures in the same conjugate image plane, making them “confocal”. In that configuration, one pinhole was placed in front of a zirconium arc light source to provide a point of light focused on the sample by an objective lens. The second objective lens focused the illuminated sample point onto the second pinhole in front of the detector. This “double focusing” system rejects the out-of-focus rays from the illuminated sample, so they do not reach the detector, which was a low-noise photomultiplier. The stage could be moved in x, y to scan the sample through the illumination point to build the resulting image. Figure 1 shows a schematic of the core optics in a modern confocal microscope, many of which remain similar to the Minsky design.
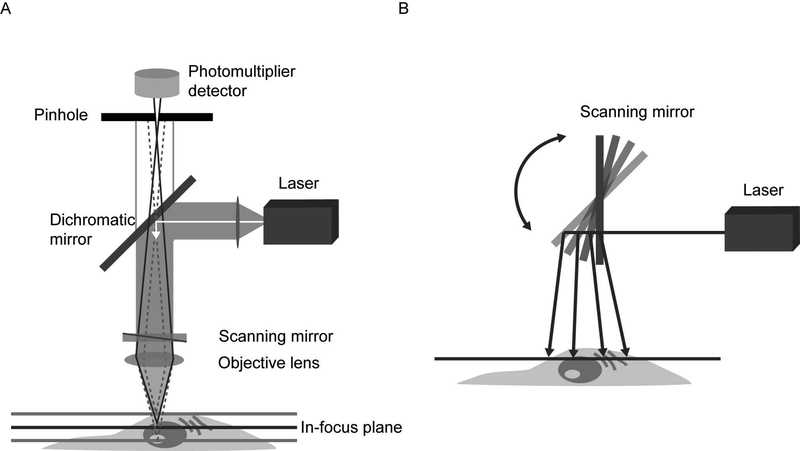
Figure 1.
Components of a confocal microscope. A. Light from a laser source is passed through collimating optics to a variable dichromatic mirror or AOBS and reflected to the objective lens which focuses the beam on a point in the sample. Scanning mirrors sweep the excitation beam over the sample point by point to build the image. Emitted fluorescence passes back through the objective lens, the dichromatic mirror or AOBS, and is detected by the PMT(s). A pinhole placed in the conjugate image plane to the focal point in the sample serves to reject out-of-focus light, which does is not picked up by the detector. In this epifluorescence configuration, the illumination and emission light both pass through the same lens, thus requiring only the detector-side pinhole. Varying the size of the pinhole changes the amount of light collected and the optical section thickness. Spectral imaging can be achieved with an array of PMTs and a diffraction grating, or prism, placed in the emission light path. B. A schematic of the scanning mirrors employed by confocal microscopes to sweep the excitation light across the sample.
The basic components of a modern confocal microscope are the pinholes, the objective lenses, and low-noise detectors in common with the original design but also typically include fast scanning mirrors, filters for wavelength selection, and laser illumination. While gas lasers (argon and helium-neon) are still in use, diode lasers, fiber lasers, and solid-state lasers are increasingly common. These light sources are more stable, more uniform, produce less heat, and emit a broad range of visible wavelengths. Detectors are still primarily highly sensitive photomultipliers (PMTs) due to the light-rejecting nature of a confocal microscope. These are essentially one spot cameras that maximize the light budget by amplifying the signal over a photoelectric device.
In light microscopy, the resolution is determined by the numerical aperture (NA) of the objective lens, the properties of the sample (index of refraction), and the wavelength of light. The lateral resolution of a confocal microscope is improved over a conventional widefield fluorescence microscope when the pinholes are closed to the minimum size providing a diffraction-limited imaging system. The best resolution that can be obtained is ~ 0.2 μm laterally and ~ 0.6 μm axially, though in practice that is not always achieved. Despite the pinholes, the axial resolution in a confocal microscope is still worse than the lateral resolution, as in widefield fluorescence microscopy. The equations used to determine lateral and axial resolution are as follows:
| Equation 1 |
| Equation 2 |
Where R is the resolution, λ is the emission light wavelength, η is the refractive index of the mounting medium (speed at which light propagates through the material), and NA is the objective’s numerical aperture. There is a tradeoff in confocal microscopy between the light collection efficiency and resolution. For dimly fluorescing samples, the pinhole may be opened to collect more light toward improving the contrast at the cost of resolution. Similarly, the resolution can be improved by closing the detection-side pinhole to a size smaller than one Airy unit at the cost of signal-to-noise. An Airy unit is defined as the zeroth order portion of the airy disc (central diffraction spot) at the image plane. At one Airy unit, the system is diffraction-limited. The theoretical considerations for confocal imaging and details on the practical use can be found in detail here (Hibbs, 2004; Pawley, 2006; Murphy & Davidson, 2012). The relative advantages and weaknesses of confocal microscopy compared with other types of fluorescence microscopy can be found in Table 2.1.2 of Combs and Shroff, 2017. For reviews on fluorescence microscopy, see Lightman & Conchello, 2005 and Inoue & Spring 1997. An example of the improvement in confocal over widefield imaging is shown in Figure 2.
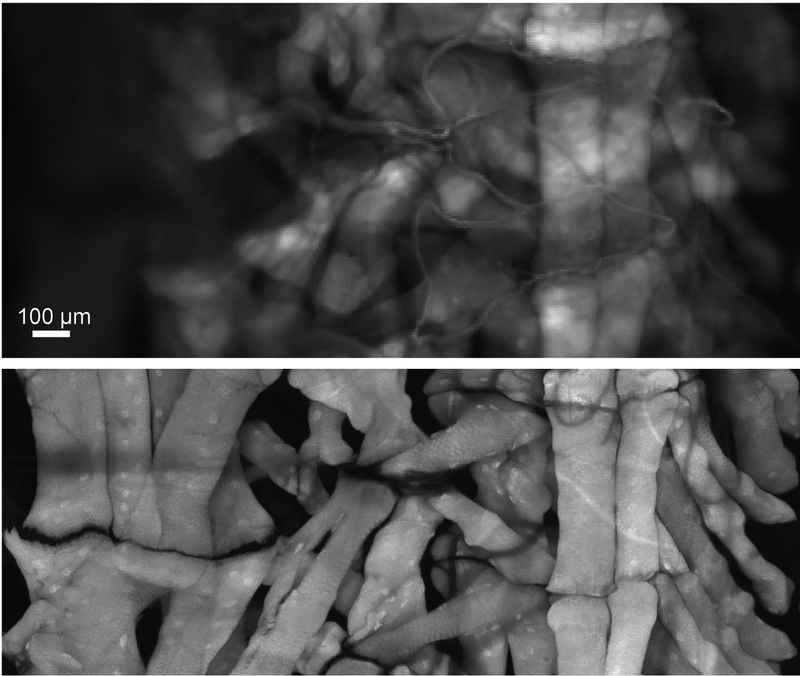
Figure 2.
Widefield vs confocal microscopy. One hemisegment of a Drosophila larval fillet stained with AlexaFluor 647-conjugated phalloidin to label the musculature. In the widefield image (top), data were collected on a widefield epifluorescence microscope. The confocal image was taken with the pinhole set to 1 Airy Unit. Both images were collected with 20x objective lenses. The confocal image required ~2 hours to build in a point scanning system and the widefield image was collected with an integration time of 1 second.
Types of Confocal Microscopes
Confocal microscopes can be distinguished by their method of scanning. The confocal image is constructed as the illumination point is moved over the sample and several strategies have been developed to accomplish that. In a stage scanning system, like the Minsky configuration, the optics are held fixed and the object is scanned by moving the microscope stage. This method has some advantages including: all points in the image have identical optical properties, edge artifacts are reduced by using only the central axis of the objective lens, and the sample size is limited only by the translation range of the stage itself. However, this requires high mechanical precision for optimal resolution, is slow compared to scanning the beam, and may lead to motion artifacts or rearrangement of tissue due to the force involved in translation. Most modern confocal microscopes scan the illumination beam across the stationary sample and are controlled with an acousto-optic tunable filter (AOTF) to rapidly turn lasers on and off, attenuate the laser power, and select the wavelength during imaging. The major classes of scanning confocal microscopes are described below.
Laser Scanning Confocal Microscopes
As the name implies, in a laser scanning confocal microscope (LSCM), a laser beam is swept over the sample by means of scanning galvanometer mirrors. Typically, the laser is directed onto a pair of scanning mirrors sweeping the beam in x and y directions of a single field of view and then moved incrementally across the entire sample to produce an image of the optical section, or slice. To collect a z-stack, the focal point is changed, and the scanning process repeated over the new slice; an example of several slices of a z-stack is shown in Figure 3. Upon collection of all optical sections from top to bottom, a 3-dimensional (3D) image can be reconstructed of the sample. The LSCM is the most common commercial implementation of this technology and can be found in most imaging laboratories with broad applications. In addition to 2D imaging of thin slices in a thick sample, these systems are often used for 3D imaging (x, y, z), and can be used for 4D imaging (x, y, z, t), and 5D imaging (x, y, z, t, λ) with spectral detectors. The principal advantages of the LSCM are the optical sectioning capability, resolution, and versatility with 3D imaging. Most systems provide multi-color imaging, the ability to adjust the pinhole size to set the optical section thickness, and region-of-interest selection. Furthermore, modern LSCM can accommodate live or fixed tissues. Important considerations include imaging speed, photodamage to the sample, and axial resolution and light penetration/collection in thick samples, which will be addressed individually further on. For detailed descriptions of these considerations, see North, 2006; Brown, 2007; and Waters, 2013.
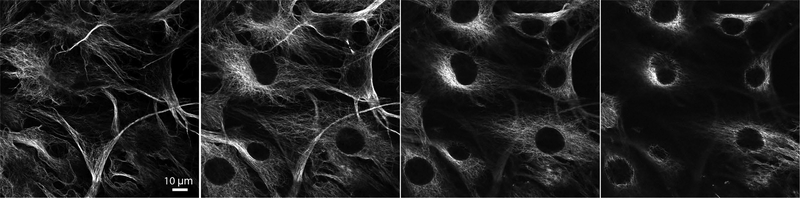
Figure 3.
Slices of a confocal imaging stack of microtubules. Hela cells were stained with anti-tubulin primary and AlexaFluor 488-conjugated secondary antibodies. Confocal z-stacks were collected at 40x magnification with an oil immersion objective and 1 μm slices.
Spinning Disk Confocal Microscopes
The LSCM is a point scanning system, where a single point is moved through the sample. There are also multi-point scanning confocal microscopes, of which the spinning disk is the oldest. Conceived in the 1880s by Paul Nipkow, the Nipkow disk is a metal disk with ~1% of the surface consisting of fixed-width holes arranged in outwardly spiraling tracks (Nipkow, 1884). These holes were positioned such that every part of the image was scanned as the disk turned and light from each point was electrically transmitted and reassembled remotely through a second disk. Decades later, an implementation of the Nipkow disk was developed for light microscopy in a tandem scanning-disk confocal microscope (Petran, et al, 1968). The confocal principle of two pinholes focusing a point of light on the sample is maintained, but the entire field can be covered at a high rate and the image is captured with a camera (CCD or EMCCD) instead of a PMT. The advantages of spinning disk confocal microscopes are the imaging speed, relatively low-light dose, and the fact that the sample does not have to be moved through the illumination. The Yokagawa implementation uses micro lenses to focus light on the pinholes to increase excitation efficiency for dimmer samples (Favro, et al, 1992). Potential drawbacks include the non-adjustable pinhole, which only comes in sizes matched to the objective lens used, artifacts from the disk alignment and synchronization of the camera speed and disk speed, and crosstalk from multiple pinholes in deeper samples.
Hybrid Scanning Confocal Microscopes
An intermediate approach between single and multi-point scanning confocal microscopes is the slit-scanning confocal, which replaces the round pinhole with a rectangular slit to reject out-of-focus light. The slit-scanning systems cover more of the sample in one field of view and significantly increase the imaging speed at the cost of rapid photobleaching and lower resolution (Sheppard & Mao, 2007). Another hybrid approach is the swept field confocal microscope (SFC), patented in 2002. The SFC can be used in a pinhole or a slit scanning mode where the apertures remain motionless while galvanometer and piezo-controlled mirrors sweep the image of the illuminated apertures across the sample. The emitted photons are directed through a complementary set of pinholes or slits onto a CCD camera. The major advantages of this approach are the speed, increase in light collection efficiency, and reduction in artifact from moving the apertures as in a spinning disk system (Castellano-Munoz, et al, 2012).
Practical Considerations
There are a number of points to consider when designing experiments for confocal microscopy, several of which are addressed below.
Objectives
Modern objective lenses have components to correct for flatness of field and chromatic aberrations, which are important for confocal microscopy, as the laser beam must pass through parts of the objective lens relatively far from the optical axis during scanning. Immersion objectives should be used to obtain the highest resolution and the refractive index should be matched to the mounting media (Dean, 1998). Most objectives are designed to be used with cover glass that has a thickness of 0.17 mm to limit artifacts due to dispersion and variable thickness. Thus, for objectives marked for 0.17 cover glass, #1.5 coverslips should be used for confocal imaging. Some objectives also come with correction collars that allow for more precise calibration to the coverslip by adjusting the spacing between elements in the objective barrel, which can help overcome immersion-related aberrations. Figure 4 shows an example of several multi-color fields of view from a sample collected with a 40x oil immersion objective. As discussed below, in addition to immersion medium and NA, the working distance of the objective lens should be noted carefully and matched to the thickness of the tissue being imaged.
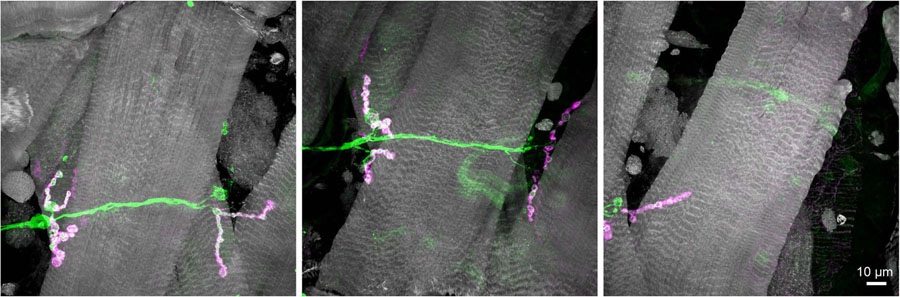
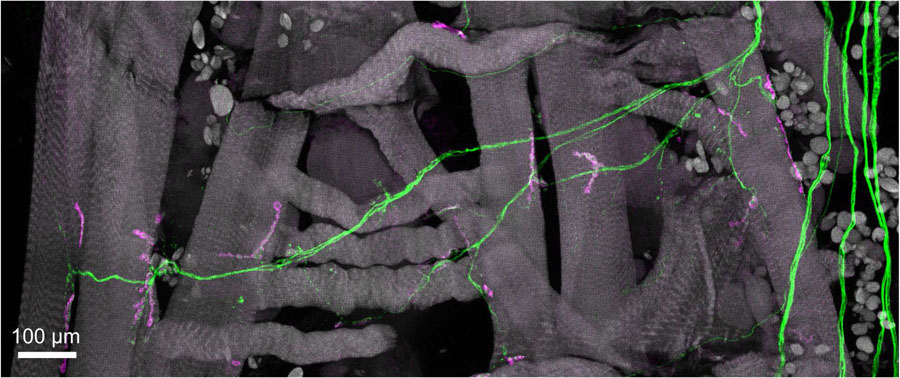
Figure 4.
Maximum intensity projection of multiple fields-of-view at 40x magnification. Three fields of view of the neuromuscular junction from a Drosophila larval fillet stained with AlexaFluor 555-conjugated phalloidin (gray), AlexaFluor 488 labeling glutamatergic motor neurons (green), and AlexaFluor 647 labeling Dlg-1 expressing type 1b post-synaptic sites. Confocal z stacks were collected with an oil immersion objective and 0.8 μm slices. Each field of view required ~10 minutes to collect with a LSCM.
Depth Penetration
Deep tissue imaging is in increasing demand and strategies to retain image quality further from the coverslip continue to emerge. Careful choice of high NA, long working distance objectives with refractive index matching to the medium can improve the penetration of a standard confocal microscope, but aberration-free imaging far from the coverslip is difficult. Recent advances in sample preparation include methods to “clear” tissue with readily available chemicals that remove lipids and other highly scattering cellular components. Several of these techniques are reviewed in Wan, et al, 2018 Multiphoton lasers are another available tool for deep imaging and are characterized by the use of pulsed near-infrared illumination that penetrates tissues deeper than visible wavelengths (Denk, Strickler, & Webb, 1990). For 3D reconstruction, the optical thickness of the sections must exceed the axial resolution of the objective lens.
Sample Size
Directly related to the depth in a confocal microscope is the overall size of the sample. The 3D capabilities of a confocal microscope can be applied to large samples by scanning the beam over a volume to collect an image stack and then moving the stage to successive fields-of-view. These “tiles” of image stacks are then stitched together either during or after the experiment by software. While this has been enabling for large tissue sections, sample thickness remains a limiting factor as discussed above. Additionally, image artifacts can arise in the process of combining component tiles and the imaging time significantly increases with size on a LSCM because the beam must still be swept over every point in the sample. An example of a multi-tile image is shown in Figure 5.
Figure 5.
Maximum-intensity projection of a multi-color, multi-tile confocal image stack. A single hemisegment of the sample in Figure 4 collected with a resonant scanning confocal microscope with a 20x air objective and 0.7 μm slices covering 4 tiles that were stitched together during image processing.
Imaging Speed
A critical consideration when planning confocal microscopy experiments is the desired acquisition speed. There are tradeoffs in confocal microscopy between imaging speed, resolution, and field-of-view. With a small field of view, as with high magnification objectives, a single field of view may be imaged faster but larger samples require more time to scan. The slit and spinning disk confocal microscopes provide a boost to the imaging speed at the cost of photobleaching. There are now confocal microscopes equipped with resonant scanners, which are fixed frequency mirrors that allow fast scanning of the sample (Figure 1B). Bidirectional resonant scanning provides a significant increase in speed for imaging very large samples, but the calibration on the mirrors must be done carefully to avoid introducing artifacts (Callamaras & Parker, 2003; Leybaert et al, 2005). As an example of imaging speed differences, Figure 5 shows a maximum-intensity projection of a couple of fields-of-view (~2 tiles) from a preparation that required 38 tiles and 109 slices to image and merge. Using bidirectional resonant scanning, a sample this size requires ~2 hours to image, and with bidirectional scanning disabled, the imaging time roughly doubles. On a LSCM that does not have resonant scanning mirrors, it takes between 6 and 8 hours to obtain a complete image. Additionally, significant increases in imaging speed typically require image averaging to obtain a reasonable signal and are far easier with very bright fluorophores. The sample in Figure 5 was collected with bidirectional scanning and averaging set at 16 images. As described below in the recent technical advances section, computational tools are being developed to reduce noise and improve the signal to noise ratio (SNR), which will hopefully reduce the requirement for image averaging in the near future.
Fluorophores
Since the introduction of the green fluorescent protein in the 1960s, numerous fluorescent proteins (FPs) have been engineered with a variety of photophysical and spectral properties, widely increasing the available palette of fluorescent probes for confocal microscopy (Shaner, Steinbach, & Tsien, 2005; Specht, Braselmann, & Palmer, 2017). Additionally, recent improvements in organic dyes have produced brighter, smaller, and more photostable products. Confocal imaging with FPs, dyes, and the wide range of available secondary antibodies has been used to great effect for determining the localization of proteins and structures in whole cells and tissues and monitoring fast dynamics in living cells. The quantum efficiency, brightness, and excitation and emission spectra should be considered when choosing the probes for any imaging experiment and the optical filters must be tuned accordingly (Ni, et al, 2017).
Sample Preparation
The mounting method for fixed tissues and the media for live samples can affect the 3D shape of the sample and the resolution that can be obtained by confocal microscopy. For live confocal imaging, best results will be achieved with media that is free from pH-indicator dyes like phenol red. When mounting fixed samples, spacers should be considered between the coverslip and the slide to prevent damaging the tissue. There are a variety of compounds for mounting fixed tissues that have different refractive indices, chemicals to increase the lifespan, slow photobleaching of the sample, etc. For details on sample preparation methods, see Smith, 2008 or Galdeen, 2011.
Recent Technical Advances in Confocal Microscopy
While the majority of confocal microscopes are based upon the Minsky principle, there have been several advances to improve their functionality. As discussed above, the spinning disk and swept field confocal microscopes, and resonant scanning mirrors increase the speed of acquiring confocal data. However, because speed is a historically limiting factor for confocal microscopy, increasing the rate of data collection continues to be a focus for technical improvements. One recent configuration is the ribbon scanning confocal microscope (Watson, et al, 2017). This functions by employing resonant scanners and a high-precision x, y stage to continuously acquire strips across the sample that are stitched together. The primary advantage of this system is the speed with which high-resolution multi-stack images can be acquired. For large, fixed samples, this technology reduces to the time to data collection for 3D stacks.
As with every microscopy technique in the last decade, there is a push to increase the resolution to enable imaging of ever smaller features. In the realm of confocal microscopy, the Airyscan technology provides 1.7x higher resolution in x, y, and z (Zeiss). The Airyscan has a 32-channel detector array with a hexagonal array of micro lenses that act as a system of very small pinholes. In this system, the primary improvement is in the signal to noise ratio (SNR) via pixel reassignment and summation of the collected images from all of the detectors. The re-scan confocal microscope (RCM) is a recently commercialized confocal technology that improves lateral resolution by 1.4 (√2). The RCM includes a re-scanning unit consisting of a pair of re-scanning mirrors between the pinhole and detector that allows for de-coupling of the magnification of the object and scanning spot (De Luca, et al, 2013). In this system, the re-scanning mirrors can be set to double the angular amplitude before directing the light to a CCD or sCMOS detector, which increases the scanning size and the apparent distance between spots. Here, the lateral resolution is independent of the pinhole, but the axial resolution is the same as in standard LSCM.
Another area of technical advance is the method of illumination. In addition to improvements in laser technology for visible wavelengths and the commercial implementation of multiphoton excitation, there are also white-light, or supercontinuum lasers. Introduced in the early 2000s, these lasers produce pulsed infrared light which is fed through a photonic crystal fiber to generate a constant energy distribution over the visible range (Engelhardt & Hoffmann, 2002; McConnell, 2005, Chiu, 2012). Selecting the individual wavelengths or wavelength ranges is accomplished with the AOTF technology mentioned above, or with an acousto-optical beam splitter (AOBS). The AOBS selects small bandlets of the spectrum by applying specific frequencies and amplitudes of acoustic waves to an appropriate crystal, which causes select colors to exit the crystal at different angles. For wavelength selection, an alternative to the AOBS is a variable bandpass filter that has spectral properties varying along the length of the filter.
Emitted light is detected in confocal microscopy with a PMT, or more recently, with a CCD camera. The highest quantum efficiency is currently provided by GaAsP detectors (gallium arsenide phosphide), a high-sensitivity PMT. The increased sensitivity permits the study of dim signals, but also provides a speed boost for brighter signals because the amount of time spent collecting the signal in each spot can be shortened. Another recent development is the hybrid detector, which is a cross between a standard PMT and an avalanche photodiode (a highly sensitive semiconductor device). They are characterized by high dynamic range, low noise, and high speeds. Additionally, arrays of the GaAsP detectors are being used for spectral imaging, where multiple bands of fluorescence emission light are collected after being split by a diffraction grating or prism (Zimmerman, Rietdorf, & Pepperkok, 2003). The increased sensitivity of the GaAsP detector is particularly important here as fewer photons are being collected after splitting.
The sum of technical advances in microscopy is driving a rapid rise in the amount of data being collected. Storing and analyzing these data is a significant challenge. However, there has been a concomitant rise in the software tools available for processing and analyzing large datasets. Commercial microscopes provide suites of tools for image registration, segmentation of features within images, algorithms for particle tracking, and many others. Additionally, freeware including ImageJ (NIH) and image processing packages for python (e.g/scikit-image, OpenCV, and others) have many built-in functions and allow for custom scripting (Schindelin, 2012). Future advances are likely to include further improvements to the computational side of confocal fluorescence microscopy and the introduction of more automated systems. Artificial intelligence (AI) and machine-learning algorithms are currently making their way into commercial software packages and many types of machine learning algorithms are available in the open source programs listed above.
Applications for Confocal Microscopy
Confocal microscopy provides the ability to collect clear images from a thin section of a thick sample with low background and minimal out-of-focus interference. Optical sectioning is a common application in the biomedical sciences and has been useful for materials science as well. In practice, a sample is put on the microscope stage and an image is collected at the top focal plane and then the stage or objective is moved up or down to the next focal plane and so on. A volumetric image or “z-stack” is the result of such an experiment and provides 3D spatial information about the sample that can be quantified (when the data is collected sub-saturation) and measurements like volume, localization, and surface area are accessible. Additionally, 3D volumes can be collected over time for 4D datasets and with multiple channels for 5D datasets.
It is increasingly common to use confocal microscopy for live imaging as well as with fixed samples. One example of a live-imaging experiment with a LSCM is shown in Figure 6, tracking muscle calcium activity during larval locomotion in Drosophila melanogaster. A number of fluorescence-based techniques are often combined with confocal microscopy including Fluorescence Resonance Energy Transfer (FRET), Fluorescence Recovery after Photobleaching (FRAP), Fluorescence Lifetime Imaging (FLIM), spectral imaging, optogenetics, and multiphoton imaging, all of which are introduced here: Paddock & Eliceiri, 2013, Murphy & Davidson, 2012.
Figure 6.
Time-series of forward larval locomotion in Drosophila melanogaster. Larvae expressing calcium biosensor GCaMP6s (Chen, 2013) in the muscles were monitored during forward locomotion in a confocal microscope at 4x magnification with the pinhole open to collect more light.
The type of confocal microscope best suited to a given application depends largely on the prioritization of imaging speed, resolution, and field-of-view – while keeping in mind photodamage to the sample. Table 1 lists the techniques outlined above with a summary of their principle, advantages and disadvantages, and experiments for that technique. Confocal microscopy can be an exceptionally quantitative technique. However, because these instruments are widely available and relatively easy to use, they are often not optimally utilized for quantitative data collection. Some examples include: a) oversampling spatially and temporally, b) photodamage in live or large fixed samples, c) mismatch of objective immersion and mounting medium, and d) overlapping fluorophores with incorrect dichroic/filters to achieve proper separation, to list a few. It is crucial in a confocal microscopy experiment to choose the correct technique, objective, fluorophores, mounting medium, and optical components to achieve the best images.
Table 1.
Comparison of confocal techniques discussed in this unit
| Principle | Advantages | Disadvantages | Application(s) | |
|---|---|---|---|---|
| Laser Scanning Confocal Microscopy (LSCM) | Rejects out-of-focus light via pinhole | Diffraction-limited resolution, versatile, optical sectioning | Phototoxicity, slow speed, axial resolution at depth | Immunohistochemistry (IHC), live imaging with bright fluorophores |
| Spinning Disk Confocal Microscopy (SDCM) | Illuminates with multiple pinholes | Diffraction-limited resolution, optical sectioning, faster than LSCM | Pinhole cross-talk, artifacts from disk-camera synchronization, fixed pinhole size, phototoxicity | Live imaging with bright fluorophores |
| Re-scan Confocal Microscopy (RCM) | Re-scans emission path onto camera with larger scan angle | √2 improved resolution, can be added to most microscope bases | Slow, photobleaching | IHC with features <200 nm |
| Airyscan | 32 detectors act as system of very small pinholes | 1.7x improved resolution in x, y, and z | Processing time, requires 32x the data of LSCM, phototoxicity | IHC with features <200 nm |
| Ribbon-scanning Confocal Microscopy | Resonant scanners with precision stage to acquire strips of images | Increased speed with very large samples | Photobleaching, processing time | IHC with very large samples |
| Resonant Scanning Confocal Microscopy | Fast mirrors sweep the beam quickly over the FOV | Increased speed | Lower SNR with ultrashort dwell times, alignment for bidirectional scan | IHC with very large samples |
| Sweptfield Confocal Microscopy (SFC) | Mirrors sweep image of illuminated apertures over sample | Increased speed | Pinhole cross-talk, phototoxicity | Live imaging with bright fluorophores |
Significance.
Confocal microscopy is widely used for fluorescence imaging in the life sciences. The last decade has seen advances in illumination sources, detectors, fluorescent probes, optics, and sample preparation techniques, which provide improvements in different combinations of speed, depth, and resolution. This paper lays out the basic principles, advantages, technical considerations, and applications for confocal microscopes to guide non-experts in determining the most appropriate confocal method for the desired experiments.
Acknowledgement
Data presented here were collected with funding from NIGMS (FI2-GM117582). This research was supported (in part) by the Intramural Research Program of the NIMH (Annual Report Number ZIA MH002800). Thanks to Louise Bertrand, Steven Ridge, and Michael Davis for assistance with data collection. The mention of any company, product, or service in this work is in no way intended as an endorsement by the National institutes of Health or the author.
Literature Cited
- Bolbat A, & Schultz C 2016. Recent developments of genetically encoded optical sensors for cell biology. Molecular Biology of the Cell. [DOI] [PubMed] [Google Scholar]
- Brown CM 2007. Fluorescence microscopy – avoiding the pitfalls. Journal of Cell Science. 120:1703–1705. [DOI] [PubMed] [Google Scholar]
- Callamaras N & Parker I 1999. Construction of a confocal microscope for real-time x-y and x-z imaging. Cell Calcium, 26(6):271–279. [DOI] [PubMed] [Google Scholar]
- Castellano-Munoz M, Peng AW, Salles FT, Ricci AJ 2012. Swept Field Laser Confocal Microscopy for Enhanced Spatial and Temporal Resolution in Live-Cell Imaging. Microscopy & Microanalysis, 18(4):753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, Looger LL, Svoboda K, Kim DS 2013. Ultra-sensitive fluorescent proteins for imaging neuronal activity. Nature, 499(7458):295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu L-D, Su L, Reichelt S, Amos WB 2012. Use of a white light supercontinuum laser for confocal interference-reflection microscopy. Journal of Microscopy, 246(2):153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean PN 1998. Confocal microscopy: Principles and Practices. Current Protocols in Cytometry, 2.7.1–2.8.12. [DOI] [PubMed] [Google Scholar]
- De Luca GMR, Breedijk RMP, Brandt RAJ, Zeelenberg CHC, de Jong BE, Timmermans W, Azar LN, Hoebe RA, Stallinga S, Manders EMM 2013. Re-scan confocal microscopy: scanning twice for better resolution. Biomedical Optics Express, 4(11): 2644–2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W, Strickler JH, and Webb WW 1990. Two-photon laser scanning fluorescence microscopy. Science 248:73–76. [DOI] [PubMed] [Google Scholar]
- Diaspro A, Bianchini P, Vicidomini G, Faretta M, Ramoino P, & Usai C 2006. Multi‐photon excitation microscopy. Biomedical Engineering Online, 5, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt J & Hoffmann J 2001. Method for scanning microscopy; and scanning microscope. U.S. Patent no. 6958858B2 (awarded 2002). [Google Scholar]
- Galdeen SA, North AJ 2011. Live Cell Fluorescence Microscopy Techniques In: Wells C, Parsons M (eds) Cell Migration. Methods in Molecular Biology (Methods and Protocols), vol 769 Humana Press. [DOI] [PubMed] [Google Scholar]
- Helmchen F & Denk W 2005. Deep tissue two-photon microscopy. Nature Methods, 2, 932–940. [DOI] [PubMed] [Google Scholar]
- Hibbs AR 2004. Confocal microscopy for biologists. New York, NY: Springer. [Google Scholar]
- Inoue S, & Spring KR 1997. Video microscopy: The fundamentals. New York, NY: Springer. [Google Scholar]
- Kreft M, Stenovec M, Zorec R 2005. Focus-drift correction in time-lapse confocal imaging. Annals of the New York Academy of Sciences. 1048(1):321–330. [DOI] [PubMed] [Google Scholar]
- Leybaert L, De Meyer A, Mabilde C, Sanderson MJ 2005. A simple and practical method to acquire geometrically correct images with resonant scanning‐based line scanning in a custom-built video-rate laser scanning microscope. Journal of Microscopy, 219(3): 133–140. [DOI] [PubMed] [Google Scholar]
- Lichtman JW & Conchello JA 2005. Fluorescence microscopy. Nature Methods, 2, 910–919. [DOI] [PubMed] [Google Scholar]
- McConnell G 2005. Noise analysis of a white-light supercontinuum light source for multiple wavelength confocal laser scanning fluorescence microscopy. Journal of Physics D: Applied Physics, 38:2620. [Google Scholar]
- Minsky M 1957. Microscopy Apparatus. U.S. Patent no. 3013467 (awarded 1961). [Google Scholar]
- Minsky M 1988. Memoir on inventing the confocal scanning microscope. Scanning 10:128–138. [Google Scholar]
- Ni M, Zhuo S, So PT, & Yu H 2017. Fluorescent probes for nanoscopy: Four categories and multiple possibilities. Journal of Biophotonics, January;10(1):11–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nipkow P 1984. German Patent no. 30105. [Google Scholar]
- Paddock SW, Eliceiri KW 2014. Laser Scanning Confocal Microscopy: History, Applications, and Related Optical Sectioning Techniques In: Paddock S (eds) Confocal Microscopy. Methods in Molecular Biology (Methods and Protocols), vol 1075 Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- Pawley JB (Ed.) 2006. Handbook of biological confocal microscopy. New York, NY: Springer. [Google Scholar]
- Petran M, Hadravsky M, Egger D, and Galambos R 1968. Tandem-scanning reflected light microscope. Journal of the Optical Society of America, 58:661–664. [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A 2012. Fiji: An open‐source platform for biological‐image analysis. Nature Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, & Tsien RY 2005. A guide to choosing fluorescent proteins. Nature Methods, 2, 905–909. [DOI] [PubMed] [Google Scholar]
- Sheppard CJR & Mao XQ 2007. Confocal microscopes with slit apertures. Journal of Modern Optics, 35(7): 1169–1185. [Google Scholar]
- Smith CL 2008. Basic confocal microscopy. Current Protocols in Molecular Biology, 14.11.1–14.11.18. [DOI] [PubMed] [Google Scholar]
- Specht EA, Braselmann E, Palmer AE 2017. A Critical and Comparative Review of Fluorescent Tools for Live-Cell Imaging. Annual Review of Physiology.79:93–117. [DOI] [PubMed] [Google Scholar]