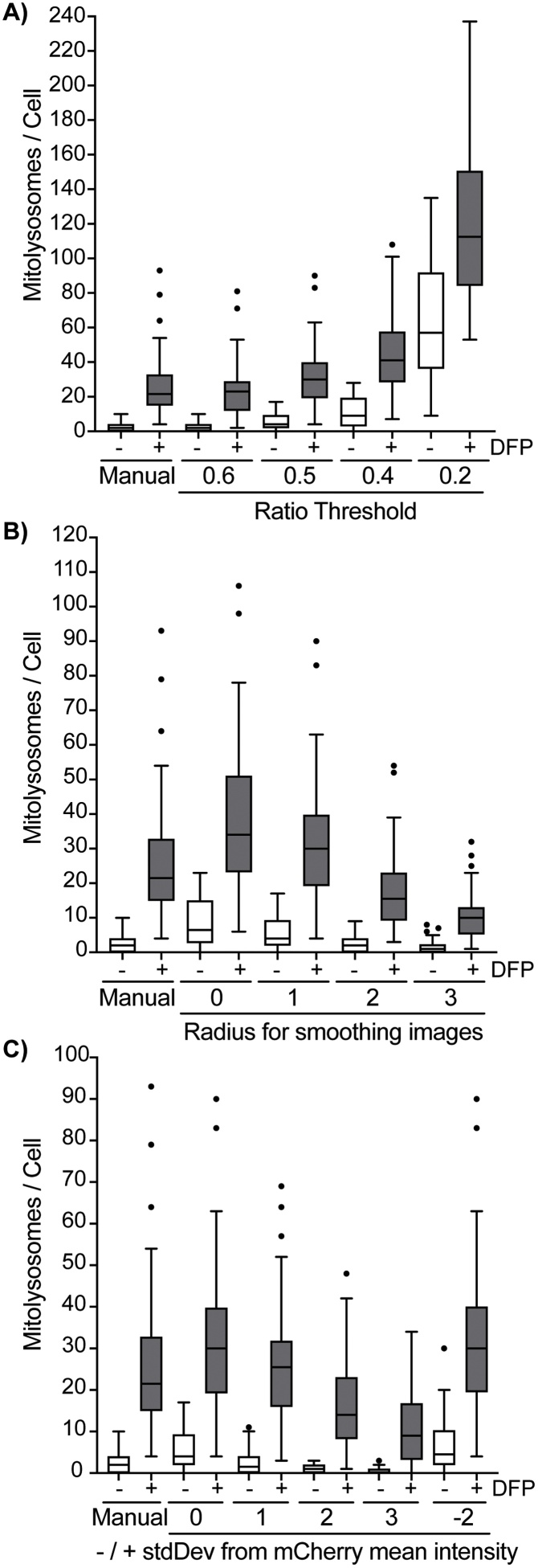
Fig. 3.
Testing mito-QC Counter thresholding settings to assess mitophagy. SH-SY5Y cells expressing the mito-QC reporter were induced with 1 mM DFP for 24 h and the number of mitolysosomes was quantified either manually or using the mito-QC Counter using 40 cells for each treatment (n = 1 experiment). (A) Boxplot assessing the effect of the Red/Green Ratio threshold in mito-QC Counter’s quantification. Cells were quantified in batch mode using a Radius for smoothing images = 1, Ratio threshold = 0.6, 0.5, 0.4 and 0.2, and a Red channel threshold = Mean + 0 stdDev. (B) Boxplot assessing the effect of the Radius for smoothing images in mito-QC Counter’s quantification. The box that depicts the value for each cell distributed from the 25th to the 75th percentiles and the median value, while the Tukey whiskers depicts the value for each cell distributed 1.5x times the interquartile range (IQR). Cells outside the whiskers range are considered outliers and are depicted as individual dots. Cells were quantified in batch mode using Radius for smoothing images = 0, 1, 2 and 3, Ratio threshold: 0.5, and a Red channel threshold: Mean + 0 stdDev. (C) Boxplot assessing the effect of the stdDev from the mean of mCherry intensity in mito-QC Counter’s quantification. Cells were quantified in batch mode using a Radius for smoothing images = 1, Ratio threshold: 0.5, and Red channel thresholds = Mean + 0, 1, 2, 3, -2 stdDev.