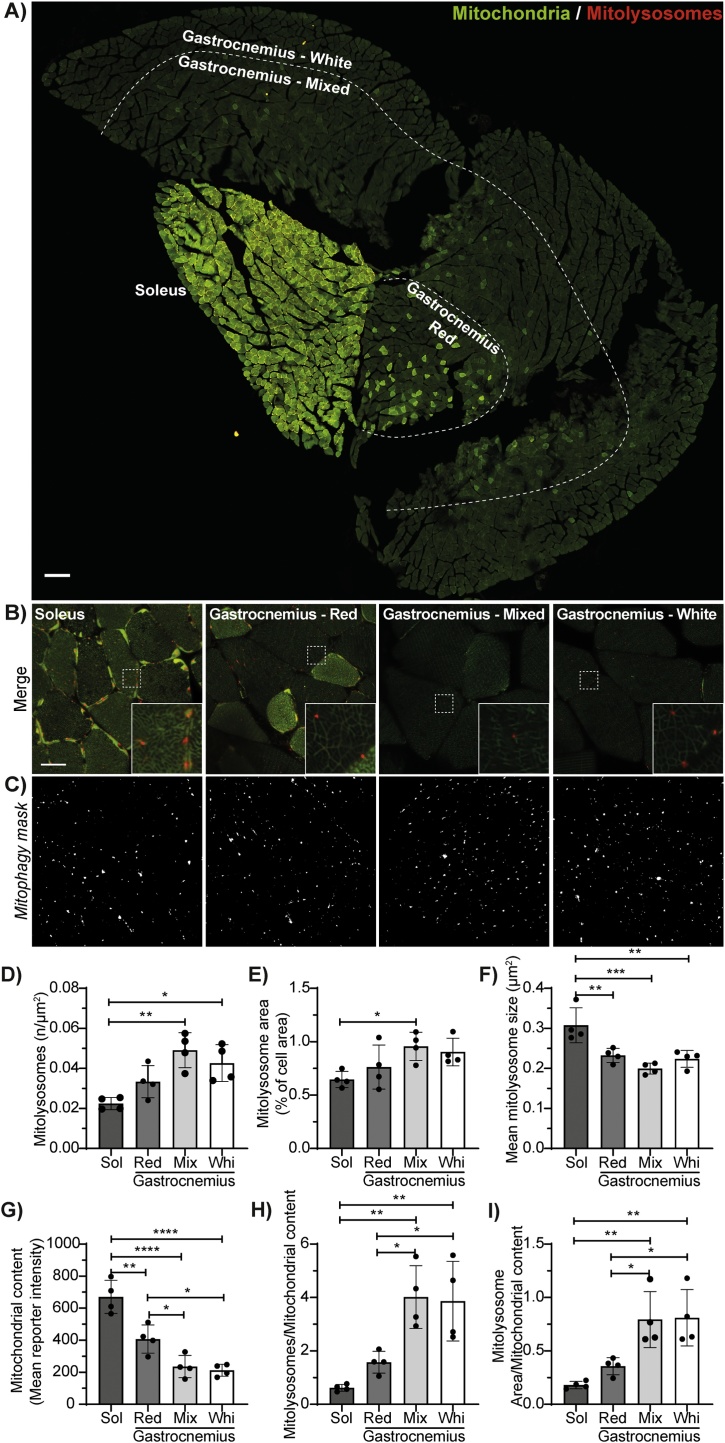
Fig. 6.
The mito-QC Counter assesses in vivo basal mitophagy in skeletal muscle. (A) Representative tile scan of a transverse section of mito-QC mouse hindlimb skeletal muscles. Scale bar =200 μm. (B) Representative images and zoomed inserts of the Soleus (Sol), the red Gastrocnemius (Red), the mixed Gastrocnemius (Mix), and the white gastrocnemius (Whi), with the (C) corresponding mitophagy channel pictures. Scale bar =20 μm. (D) Number of mitolysosomes per tissue area. (E) Mitolysosome area expressed in percent of the tissue area. (F) Mean size of the mitolysosomes. (G) Mitochondrial content estimated with the mean intensity per pixel of the mito-QC reporter. (H) Number of mitolysosomes per mitochondrial content. (I) Mitolysosome area per mitochondrial content. Quantification was performed with the following parameters: Radius for smoothing images = 1, Ratio threshold = 1, and Red channel threshold = mean + 1 stdDev. Statistics were realised using one-way ANOVAs followed by a Tukey post-hoc test. n = 4 mice. Statistical significance is displayed as *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).