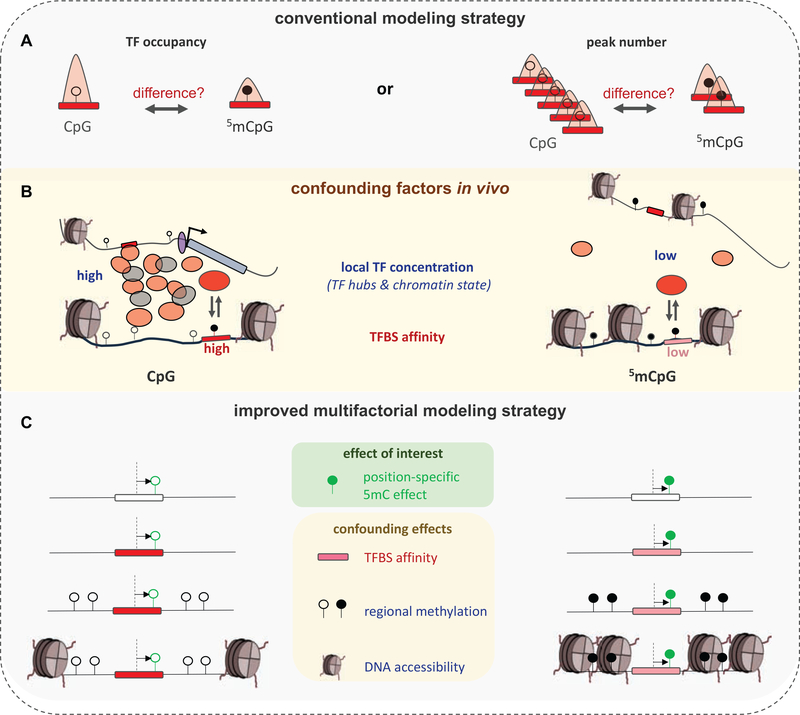
Figure 4: The challenge of quantifying the effect of cytosine methylation on TF binding in vivo.
(A) Conventional approaches typically consider whole-genome bisulfite sequencing (WGBS) and TF occupancy data (ChIP-seq) in conjunction to infer TF binding preferences for methylated binding sites in vivo. Two approaches are shown that either compare the ChIP-seq signal at unmethylated and methylated CpG containing ChIP-seq peaks (top-left), or the total overlap between WGBS sites and TF peaks split by CpG methylation status (top-right). (B) Confounding features that influence TF occupancy (IP signal) at any given site are typically not considered in conventional analysis of in vivo data. These include (i) variation in local TF concentration (likely the result of transient, phase-separated TF hub assemblies), and (ii) the affinity of each individual binding site. (C) To isolate the effect of individual methylation marks from among these confounding site-specific factors, conventional models need to be updated. Local TF concentration cannot be estimated directly, but features such as DNA accessibility or regional methylation level can be used to control for its variation.