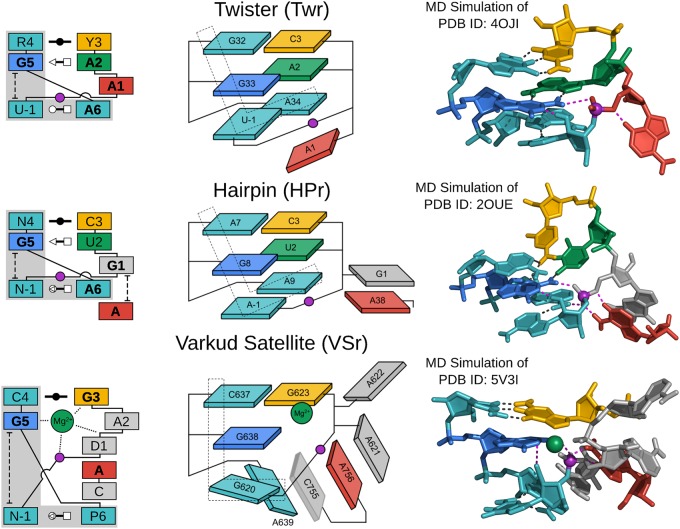
FIGURE 2.
G + A paradigm. Symbolic secondary structure (left), cartoon-block schematic (middle), and 3D atomic (right) representations of the specific L-platform/L-scaffold composite motif for ribozymes of the “G + A” paradigm (Twr, HPr, and VSr), categorized as such with a guanine (G in blue) and adenine (A in red) implicated as the general base and acid, respectively. Generalized nucleotide residue numbering is used for the secondary structure (left) as in Figure 1, whereas the cartoon-block schematic (middle) uses the canonical residue numbering of the specific ribozyme being illustrated. Base-pair symbols (Leontis and Westhof 2001; Leontis et al. 2002) are described in the Notation section, and the color scheme is the same as in Figure 1: general base (G5) in blue, stacking/pairing “L” residues in teal, L-anchor in green, L-pocket nucleobase in yellow, and the scissile phosphate in magenta. Structural h-bonds are in black and h-bonds implicated in the catalytic mechanism are shown in magenta. The active site metal ion in VSr that plays an organizational role as the L-anchor is shown in green. Bold font is used for residues conserved with a frequency greater than 97% for ribozymes where consensus sequence information is available, as well as residues that are otherwise critical for activity. 3D atomic representations derived from MD simulations of each ribozyme: Twr (Gaines et al. 2019), HPr (Heldenbrand et al. 2014), and VSr (Ganguly et al. 2019a).