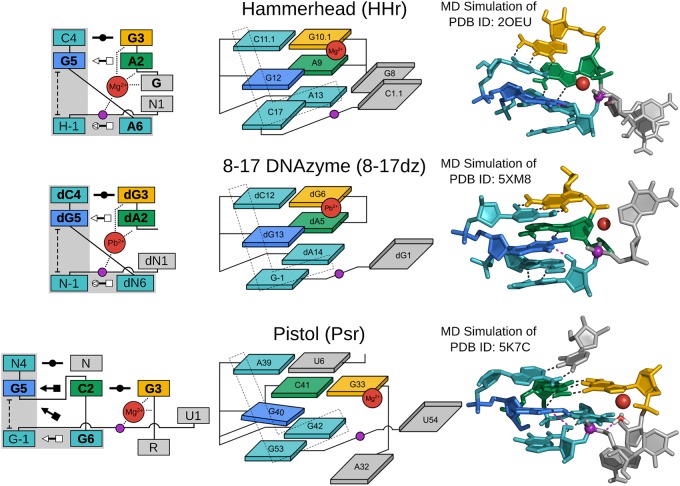
FIGURE 3.
G + M+ paradigm. Symbolic secondary structure (left), cartoon-block schematic (middle), and 3D atomic (right) representations of the specific L-platform/L-scaffold composite motif for ribozymes and DNAzyme of the “G + M+” paradigm (HHr, 8–17dz, and Psr), categorized as such with a guanine (G in blue) and a metal ion (M+ in red) implicated as the general base and acid, respectively. Generalized nucleotide residue numbering is used for the secondary structure (left) as in Figure 1, whereas the cartoon-block schematic (middle) uses the canonical residue numbering of the specific ribozyme/DNAzyme being illustrated. Base-pair symbols (Leontis and Westhof 2001; Leontis et al. 2002) are described in the Notation section, and the color scheme is the same as in Figure 1: general base (G5/dG5) in blue, stacking/pairing “L” residues in teal, L-anchor in green, L-pocket nucleobase in yellow, and the scissile phosphate in magenta. Structural h-bonds are in black and h-bonds implicated in the catalytic mechanism are shown in magenta. The active site metal ion that plays a role in the chemical steps of the reaction is shown in red. Bold font is used for residues conserved with a frequency greater than 97% for ribozymes where consensus sequence information is available, as well as residues that are otherwise critical for activity. 3D atomic representations derived from MD simulations of each ribozyme: HHr (Lee et al. 2013; Chen et al. 2017), 8–17dz (Ekesan and York 2019), and Psr (Kostenbader and York 2019).