Figure 2.
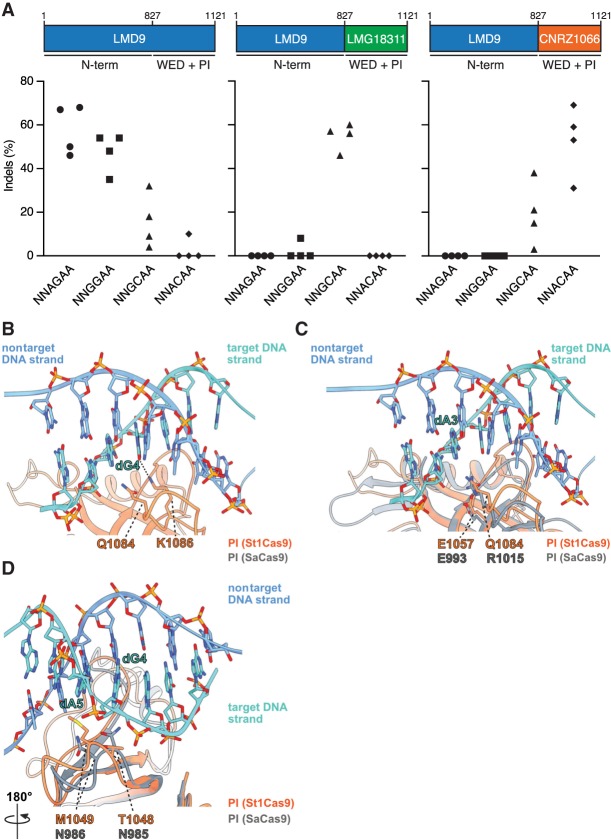
Structural basis for PAM specificity of engineered St1Cas9 variants with expanded targeting range. (A) Schematic representation of St1Cas9 hybrid proteins containing the N terminus of LMD9 and the C-terminal domains (WED + PI) of LMG18311 or CNRZ1066. To determine the activity of St1Cas9 variants programmed with sgRNAs compatible with different PAMs, K562 cells were transiently transfected with single-vector constructs driving expression of St1Cas9 and its sgRNA. For each PAM and nuclease combination, four different sgRNAs (targets) were tested. Surveyor assays were performed 3 d later to determine the frequency of indels. An expression vector encoding EGFP (−) was used as a negative control. The experiment was performed twice and yielded equivalent results; only one is shown. (B) Close-up view of the 5′-GCAGAAA-3′ PAM bound to the St1Cas9 (DGCC7010) PI domain (PDB: 6RJD). The target (turquoise) and nontarget (blue) strands are shown as sticks (the phosphate–sugar backbones are also shown as ribbons). The ribbon representation of the PI domain is orange. The hydrogen bonds between the side chain of St1Cas9 K1086 and the nucleobase of dG4 is shown as a dashed line. (C,D) The PI domains of St1Cas9 and SaCas9 (PDB: 5CZZ, gray ribbon) are superimposed. (C) The St1Cas9 Q1084 and SaCas9 R1015 occupy the same positions relative to the PAM (dA3). The St1Cas9 E1057 and SaCas9 E993 occupy the same positions relative to St1Cas9 Q1084 and SaCas9 R1015, respectively. (D) St1Cas9 T1048 and M1049 (substituted for N1048 and D1049 in some variants) superimpose onto SaCas9 N985 and N986 that specify purines in positions 4 and 5 of the PAM. See also Supplemental Figure S3.