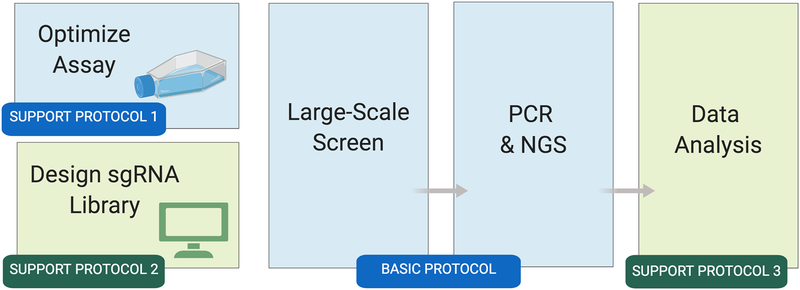
Figure 1: Overview of the Drosophila cell CRISPR pooled screen workflow.
Wet-bench activities are shown in blue; bioinformatics steps are shown in green. Once the cell-based assay is optimized (Support Protocol 1) and a library is obtained or built (Support Protocol 2), the large-scale screen can begin. The Basic Protocol takes about 45 days to complete. After the screen, the resulting next-generation sequencing data are analyzed to identify screen ‘hits’ (positive results) at the reagent and gene levels (Support Protocol 3).