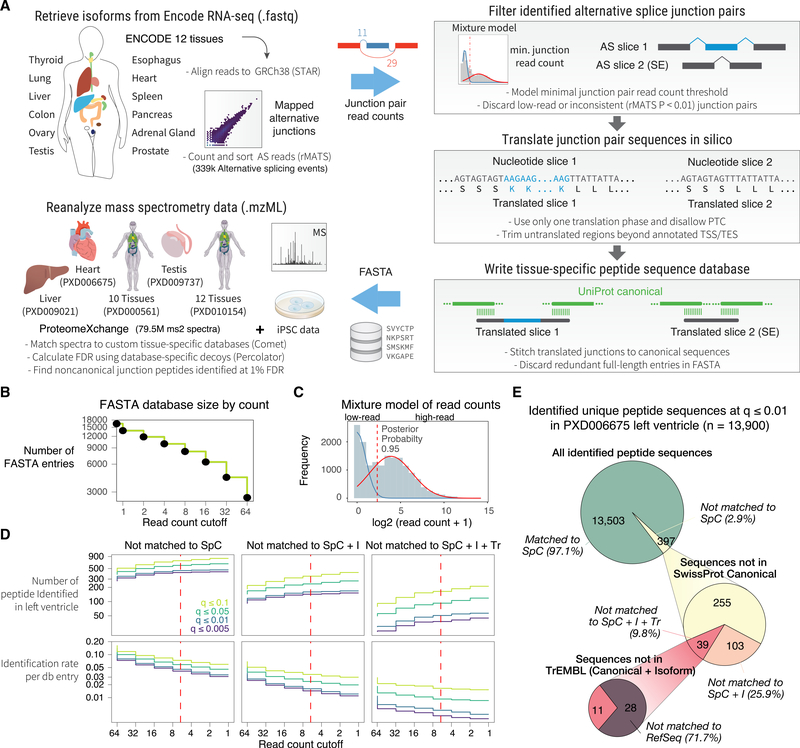
Figure 1. Splice-Junction-Centric Approach to Identify Protein Isoforms.
(A) Schematic of the method. ENCODE RNA-seq data from 12 human tissues are mapped to GRCh38. AS pairs are extracted then filtered by junction read counts and consistency. Candidate junctions are trimmed using Ensembl GTF-annotated translation start site (TSS) and translation end site (TES) and then translated in-frame by using either GTF-annotated reading frames or by choosing a frame that does not lead to PTC. The translated junction pairs are extended to encompass the full protein sequence. The created custom tissue-specific databases are used to identify noncanonical protein isoforms in public and original MS data.
(B) Number of translated sequences versus minimal skipped junction read count threshold following in silico translation in ENCODE human heart data. Inclusion of low-read junctions increases database size.
(C) Gaussian mixture fitting overlaid on skipped junction read counts of all AS events in the heart database. Dotted line: chosen threshold.
(D) Number of identified noncanonical isoform sequences in the reanalyzed human heart left ventricle MS data versus junction read count thresholds. Color: Percolator FDR cutoff calculated with database-specific decoys.
(E) Proportion of identified distinct peptide sequences in the left ventricle dataset (13,900 total) not matchable to SwissProt canonical (SpC), SwissProt canonical + isoform (SpC + I), TrEMBL (Tr), or RefSeq.