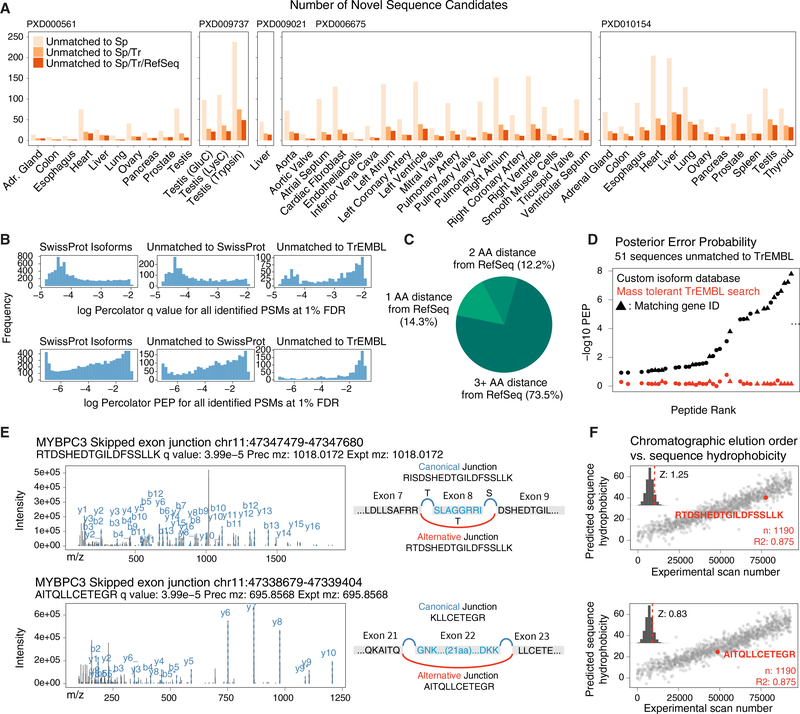
Figure 4. Splice Junctions Include Peptides Undocumented in Common Databases.
(A) Number of undocumented sequence candidates in each reanalyzed tissue across 5 public human proteome datasets.
(B) Distribution of Percolator FDR and posterior error probability (PEP) of noncanonical sequences that are matched to SwissProt isoforms (left) against those not in SwissProt (middle) or TrEMBL (right).
(C) Proportion of peptide sequences that are not mappable to RefSeq, allowing 1, 2, or 3 mismatches.
(D) Comparison of −log10 Percolator PEP for 51 left ventricle peptide spectrum matches to sequences not in TrEMBL versus the results from the corresponding spectra in a mass tolerant open search against TrEMBL.
(E) Tandem mass spectra of two identified splice junction peptides (RTDSHEDTGILDFSSLLK and AITQLLCETEGR for MYPBC3) not found in SwissProt, TrEMBL, or RefSeq.
(F) The predicted hydrophobicity of the two undocumented sequences shows the sequence eluted at the expected retention time when the spectrum was acquired. Inset: Z score of residuals from best-fit line.