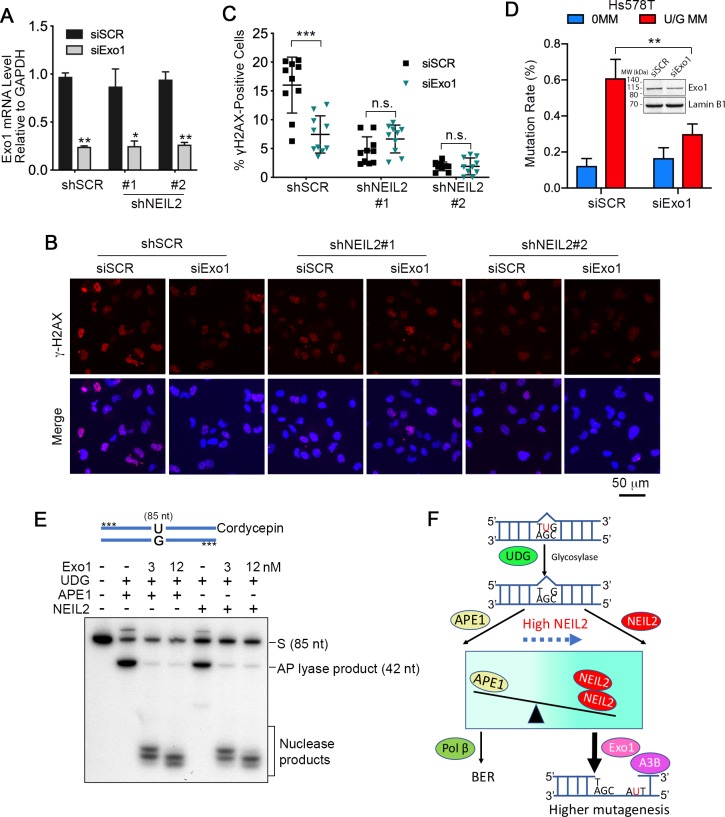
Figure 5. Exo1 generates single-stranded substrates vulnerable to A3B and DNA damage from NEIL2 products.
(A) qRT-PCR analysis of siRNA knockdown of Exo1 in NEIL2-stable-knockdown Hs578T cell lines (shNEIL2#1 and shNEIL2#2). shNEL2#1 targets NEIL2 3’UTR; shNEIL2#2 targets NEIL2 ORF; shSCR, scramble shRNA; siSCR, scramble siRNA. Error bars represent s.d., n = 3. *P < 0.05; **P < 0.01 by two-tailed unpaired Student’s t test. (B) Immunostaining of γH2AX foci in Hs578T cells depleted of Exo1 by siRNA in NEIL2-stable-knockdown Hs578T cell lines. A3B-3HA was expressed in these cell lines for 48 h before immunostaining. Scale bar, 50 μm. (C) Quantification of the percentage of cells with γH2AX foci from (B). Data are represented as mean ± s.d. (n=10 randomly selected microscopic fields). ***P < 0.001; n.s., no significant difference by two-tailed unpaired Student’s t test. (D) Effect of Exo1 depletion on U/G MM repair-induced mutation rate in Hs578T cells. Insert, western blot analysis of Exo1 knockdown efficiency by siExo1 (20 nM siRNA). Lamin B1 serves as a loading control. 0 MM, no mismatch; U/G MM, U/G mismatch. **P < 0.01 by two-tailed unpaired Student’s t test. (E) APE1 and NEIL2 scission products serve equally well as Exo1 substrates. 5’-phosphorothioate-modified (denoted by asterisk) U-containing oligo (85 nt) was 3’-labeled with cordycepin and used as the substrate for UDG, APE1 or NEIL2, and Exo1(human Exo1 protein). Nuclease products are bracketed. (F) Model depicting NEIL2-diversion of BER to Exo1-mediated resection to generate single-stranded A3B substrate.