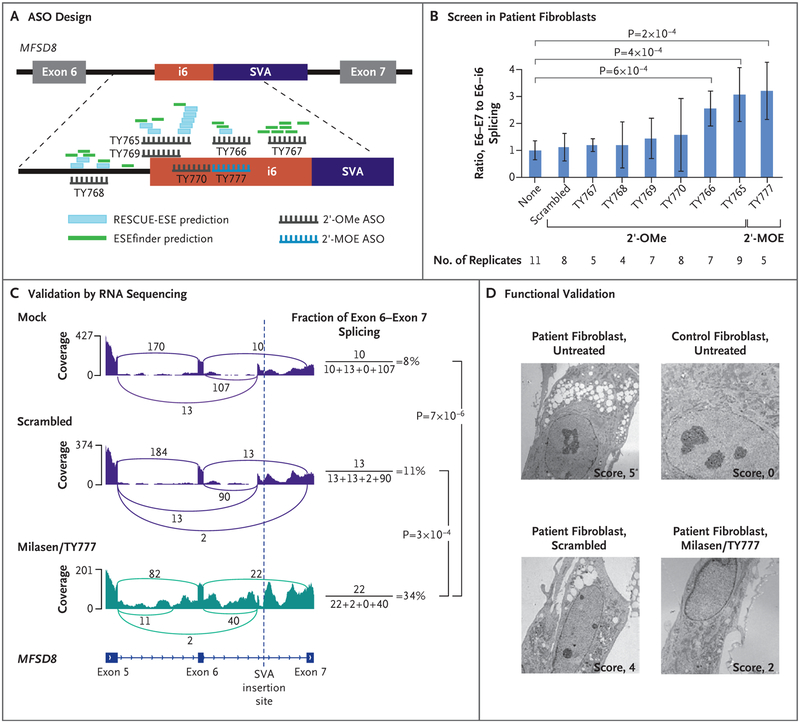
Figure 2. Antisense Oligonucleotide Drug Development.
Panel A shows the location and chemistry of the ASOs that were designed to block the i6.SA splice acceptor site or exonic splice enhancer (ESE) elements. (Additional details are provided in Table S1.) The ESE elements were predicted with RESCUE-ESE and ESEfinder.14,15 2′-MOE denotes 2′-O-methoxyethyl, and 2′-OMe 2′-O-methyl. Panel B shows the ratio of the normal exon 6–exon 7 (E6–E7) splicing to the abnormal exon 6–intron 6 (E6–i6) splicing (normalized to a no-transfection control), measured in patient fibroblasts that were transfected (for 24 hours at 100 nmol per liter) as indicated. To measure splice isoform-specific levels, multiplex reverse-transcriptase polymerase chain reactions were conducted with isoform-specific primer sets, and then the intensity of the isoform-specific bands was quantified by gel electrophoresis (Fig. S6). “Scrambled” indicates a nontargeting oligonucleotide (TY772). I bars indicate 95% confidence intervals of the means. P values were calculated by two-sided t-test. Panel C shows RNA sequencing (RNA-seq) analysis validation of the splice-correcting effect of milasen (TY777). For the calculation of the fraction of normal splicing (exon 6–exon 7), three other splicing events that are mutually exclusive with the normal splicing were considered. Splicing events supported by only one read are not shown. P values were calculated by Fisher’s exact test. Panel D shows intracellular vacuoles, visualized by electron microscopy, in control fibroblasts (MFSD8 wild-type human foreskin fibroblast; BJ cell line) and in patient fibroblasts that are either untreated or transfected with the indicated oligonucleotide. Scoring was performed on a scale of 0 to 5, with 0 representing the lowest and 5 representing the highest level of vacuole accumulation.