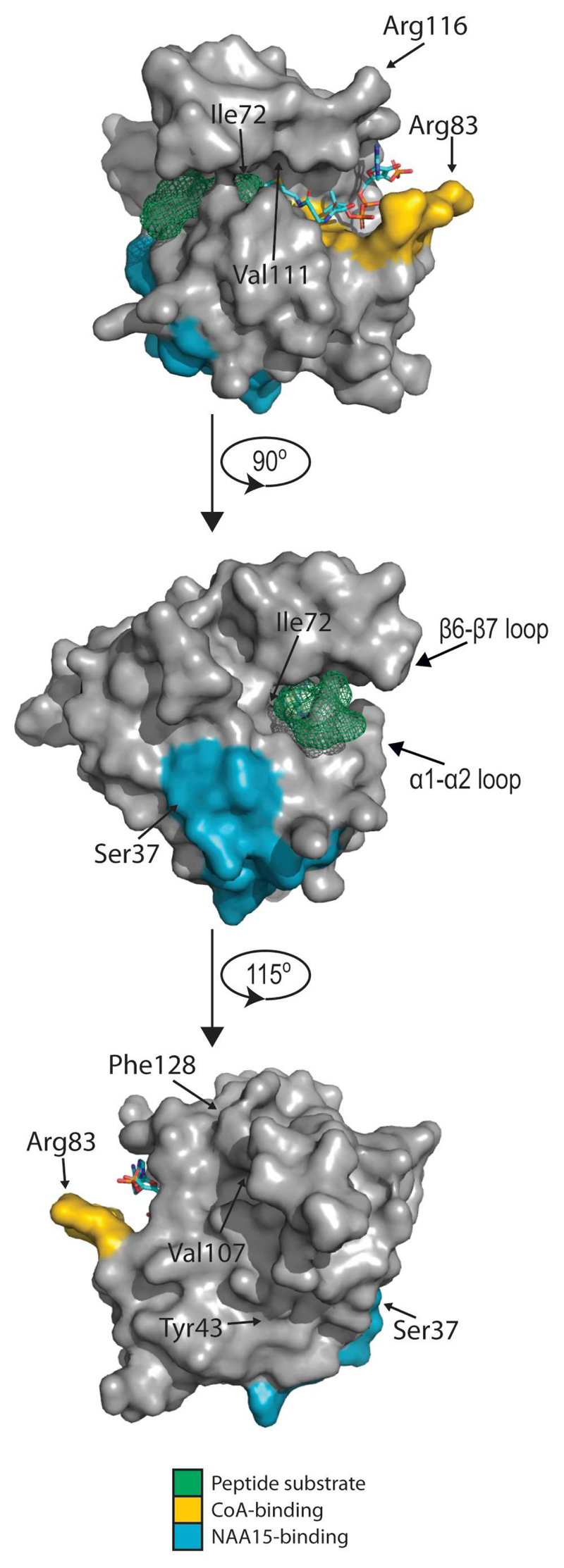
Figure 6. Structure of NAA10 and location of pathological mutation sites.
A structural model of NAA10 with SASE peptide substrate and CoA was made in PyMol by aligning the structure of human NAA10 in complex with NAA15 (not shown) (PDB ID: 6C9M; (Gottlieb and Marmorstein, 2018)) to the structure of Schizosaccharomyces pombe NAA10 solved in complex with NAA15 (not shown), CoA and substrate peptide (4KVM; (Liszczak et al., 2013)). CoA is shown as a licorice model colored according to element, SASE substrate peptide is represented in green, CoA-binding residues in yellow and NAA15-binding residues in blue. Mutation sites are indicated with arrows.