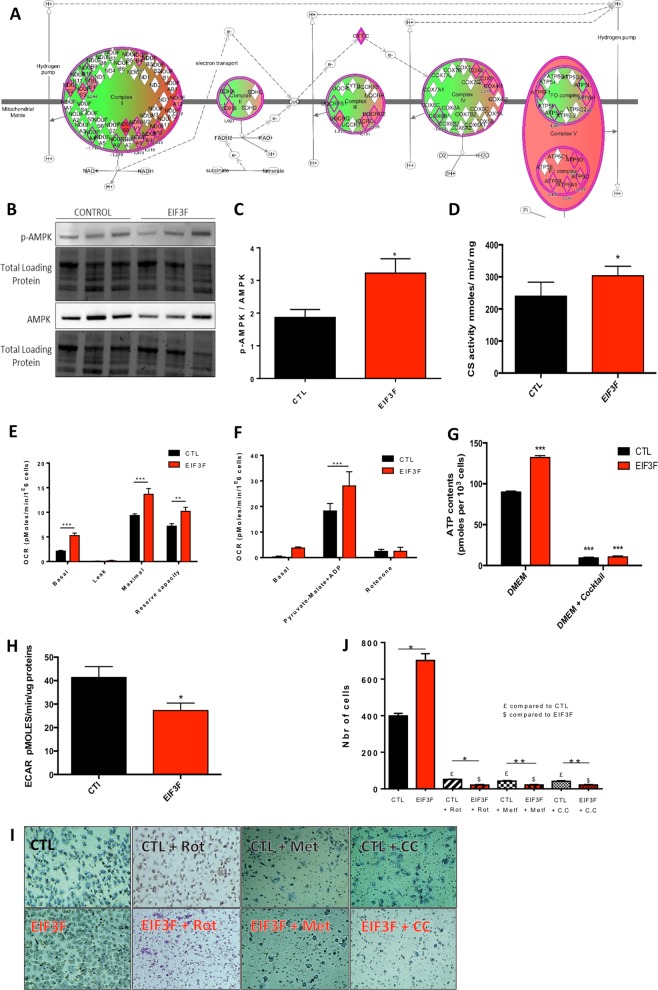
Fig. 5.
EIF3F overexpression promotes an oxidative shift in human lung adenocarcinoma A549 cells. a The mitochondrial proteins involved in energy metabolism and upregulated in the proteome of EIF3F-A549 mice orthotopic lung tumors are shown in red in the respiratory chain schematic representation. b Representative and c quantitative expression level of p-AMPk and total AMPK (N = 8). d Citrate synthase enzymatic activity measurement (N = 3) indicative of the cellular mitochondrial content in A549 and EIF3F-A549 cells. e Mitochondrial respiration measured in different respiratory states: basal (DMEM 1 g/L), leak (oligomycin), maximal (CCCP); see ‘Methods’. The reserve capacity was calculated as the difference between the maximal and the basal respiratory rates. The different rates were corrected for nonmitochondrial respiration determined after potassium cyanide addition (N = 7). f Mitochondrial respiration measurement in permeabilized cells (A549 and EIF3F-A549) in presence of pyruvate-malate as energy substrate. g Steady-state ATP content in A549 and A549-EIF3F cells. The part of ATP produced by mitochondria was inhibited using a cocktail of OXPHOS inhibitors (see ‘Methods’) (N = 8). h Extracellular acidification rate of A549 and A549-EIF3F cells (N = 5). i Quantification, and j representative images of cell migration (transwell) assay performed in CTL-A549 and EIF3F-A549 cells using compound C (5 μM), an AMPK inhibitor and metformin (5 mM), a complex I inhibitor (N = 3). For each condition the respective treated (or untreated) control was used. *P < 0.05, **P < 0.01, ***P < 0.001