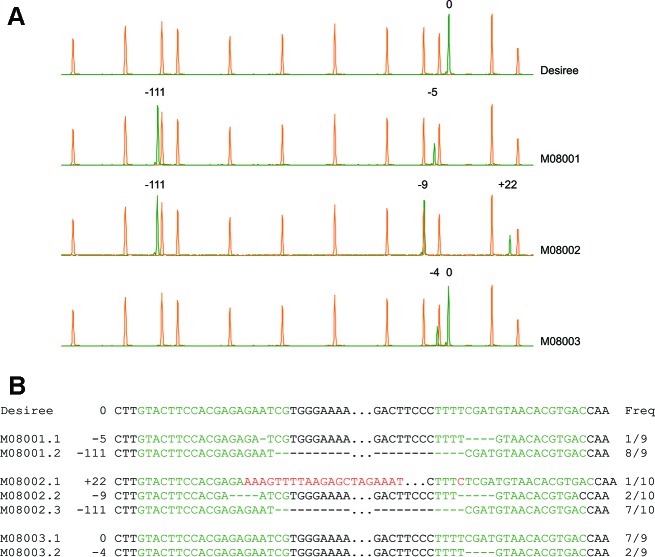
Figure 2.
Identification of edited lines using High Resolution Fragment Analysis (HRFA) and characterization of mutations by sequencing (A) Electropherograms of HRFA obtained for wild type Desiree and lines M08001, M08002, and M08003. The orange peaks correspond to the elution points of the size standard and green peaks correspond to elution of the StPPO2 gene fragments. The elution of the wild type fragment is set to 0 and the number of bases inserted (+) or deleted (−) in each fragment is indicated above the respective peak (B) Sequencing of a partial fragment of the StPPO2 alleles in selected lines. Target sites for the sgRNAs are marked in green letters. Deleted nucleotides are indicated as hyphens and inserted bases are marked in red letters. The frequencies obtained during Sanger analysis are indicated, as the number of clones carrying each allelic variant related to the total number of sequenced clones.