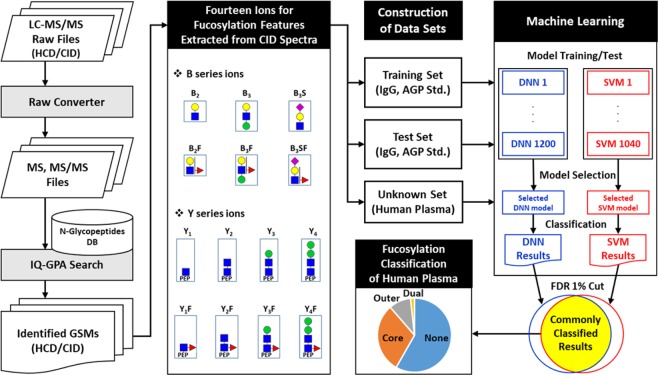
Figure 1.
The computational workflow for classifying the fucosylation of N-glycopeptides using machine learning. The relative intensities of 14 fucosylation features extracted from CID tandem MS spectra of identified N-glycopeptides were calculated and used to classify fucosylation using the DNN and SVM. Training and testing data sets were constructed with N-glycopeptides identified from standard IgG and AGP glycoproteins using IQ-GPA. The DNN and SVM models were constructed with TensorFlow (ver. 0.12.0) and the R package e1071 (ver. 3.4.3), respectively. The best-performing model was selected from each machine learning method, and classified N-glycopeptides were filtered with <1% FDR using a random decoy. Finally, the DNN and SVM were used to classify an unknown data set from human plasma according to four types of fucosylation: none, core, outer, and dual. Green circles = nomannose; yellow circles = angalactose; blue squares = N-acetylglucosamine; red triangles = fucose; and pink diamonds = N-acetylneuraminic acid.