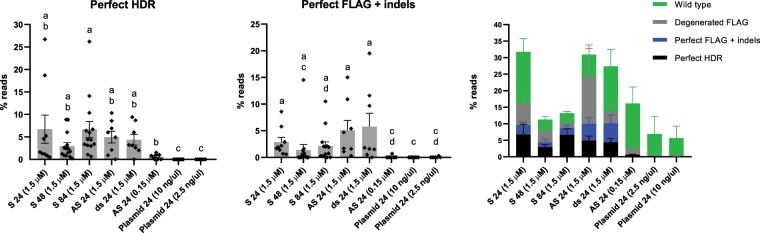
Figure 2.
NGS results. A fragment covering the entire CRISPR target site was amplified (76 fish) prior to Illumina MiSeq sequencing. When reporting read counts with the inserted sequence, we distinguished between the following groups; (a) Perfect HDR (reads with a perfect match to the entire target sequence), and (b) Perfect FLAG + indels (reads with a correct insert sequence but mismatches/indels in the homology arms). We also reported reads with mismatches in the insert sequence (referred to as degenerated FLAG) and wild type reads (Supplementary Fig. 3). All the data are summarized in (c). Read counts for each group are given in % of the total number of reads with at least 100 identical reads, for each sample. Individual samples are represented by black diamonds, and grouped for each of the different repair templates, at different concentrations (represented by grey bars). The error bars indicate the SEM of the mean for each group. Non-parametric statistics (Kruskall-Wallis) were performed to analyze the differences in HDR efficiencies between the different repair templates. Different lower-case letters indicate significant differences (P < 0.05).