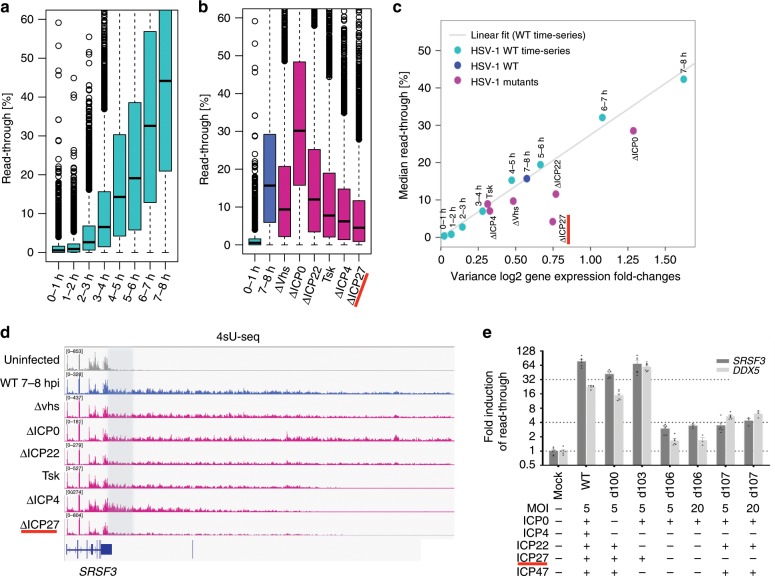
Fig. 1. ICP27 is a major contributor for HSV-1-induced disruption of transcription termination.
a Boxplots showing the distribution of read-through at different time points post-infection during HSV-1 wild-type infection (time-course). b Boxplots showing the distribution of read-through in cells infected with wild-type and various mutant viruses at 7–8 h (WT, Δvhs, ΔICP4, ΔICP27) and 11–12 h (ΔICP22, ΔICP0) post-infection. Please note that read-through at 7–8 h post-infection shown here is from a repeat experiment performed together with the mutant viruses. Read-through at 0–1 h post-infection from the WT time-course is also shown. c Median read-through values for each condition and time-point are plotted against the variance in log2 gene expression (=gene FPKM) fold-changes. The gray curve indicates the result of linear fit on all HSV-1 infection time-points from the time-course experiment. d Mapped 4sU-seq reads for SRSF3 in cells infected with wild-type or various mutant HSV-1 strains. The region where transcription termination occurs in mock-infected cells is shaded. e Primary human fibroblasts infected were infected for 8 h with mutant viruses lacking various immediate early genes. The genes that are still expressed by the individual mutants as well as the employed multiplicity of infection are indicated in the table below the graph. Read-through transcription was quantified by qRT-PCR data and plotted as mean ± s.e.m. (n = 5).