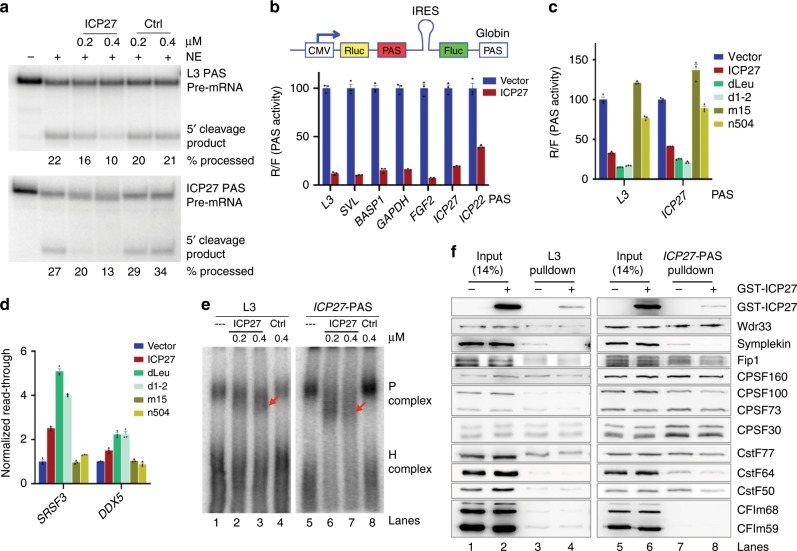
Fig. 4. ICP27 inhibits mRNA 3′ processing.
a In vitro cleavage assay with HeLa nuclear extract (NE) using L3 (top panel) or ICP27 (lower panel) PAS pre-mRNA substrate. 5′ cleavage product is marked. Different amounts of recombinant MBP-ICP27 (labeled as ICP27) or MBP-MS2 (Ctrl) were added to NE. b Dual luciferase reporter assay. The top panel is a diagram for pPASPORT vector and the site where the PAS to be tested is inserted is marked as PAS (in a red box). Seven distinct PASs were cloned into pPASPORT, co-transfected with a vector or ICP27-expression plasmid. The Rluc/Fluc ratio, which measures cleavage/polyadenylation efficiency at the test PAS, are plotted as mean ± s.d. c pPASPORT assays with L3 and ICP27 PAS. Empty vector, ICP27 or its various mutants were co-expressed with the pPASPORT constructs and renilla/firefly luciferase ratio was plotted as mean ± s.d. d Empty vector, ICP27 or its various mutants were transiently expressed and transcription read-through at the SRSF3 and DDX5 genes were measured by qRT-PCR and plotted as mean ± s.d. e In vitro cleavage/polyadenylation reactions (similar to a) were resolved on a native gel and visualized by phosphorimaging. The mRNA 3′ processing complex (P complex) and the heterogeneous complex (H complex) are marked. f 3MS2-L3 or 3MS2-ICP27 PAS RNA substrates are incubated under cleavage/polyadenylation conditions and the assembled RNP complexes were pulled down by using amylose beads and analyzed by western blotting. Source data are provided as a Source Data file.