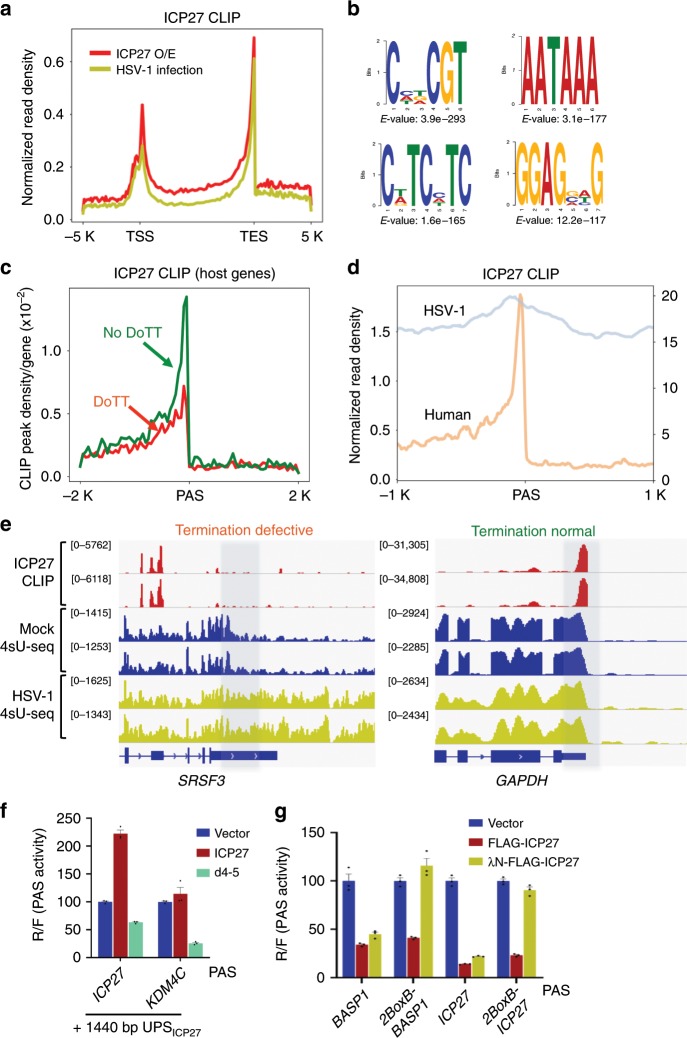
Fig. 6. The bimodal activities of ICP27 are regulated by ICP27–RNA interactions.
a Global distribution of ICP27 CLIP-seq signals along cellular genes. TSS: transcript start site. TES: transcript end site. b ICP27 CLIP-seq peak regions were analyzed by DREME and examples of the top enriched motifs are shown. c ICP27 CLIP-seq peak densities at PAS of host genes with DoTT (red) and without DoTT (green). d ICP27 CLIP-seq peak densities at the PASs of HSV-1 and host genes. e ICP27 CLIP-seq tracks at SRSF3 and GAPDH genes. For comparison, 4sU-seq tracks in mock- and HSV-1-infected cells were also shown. The region where termination normally occurs is shaded. f Wild-type or d4-5 (ΔRGG) mutant ICP27 or control (empty vector) were co-expressed with pPASPORT constructs containing ICP27 or KDM4C PAS fused to the ICP27 UPS. PAS activities (Renilla/firefly ratio) were plotted as mean ± s.d. g Protein tethering assay. Two tandem copies of BoxB hairpin are inserted upstream of the PASs. λN-tagged proteins will be tethered to RNAs via interactions with BoxB. pPASPORT constructs containing PASs labeled on the x axis were co-transfected with vector or Flag-ICP27 or λN-Flag-ICP27 expressing plasmids. mRNA 3′ processing efficiencies (Rluc/Fluc) at the PAS are plotted on the y axis as mean ± s.d.