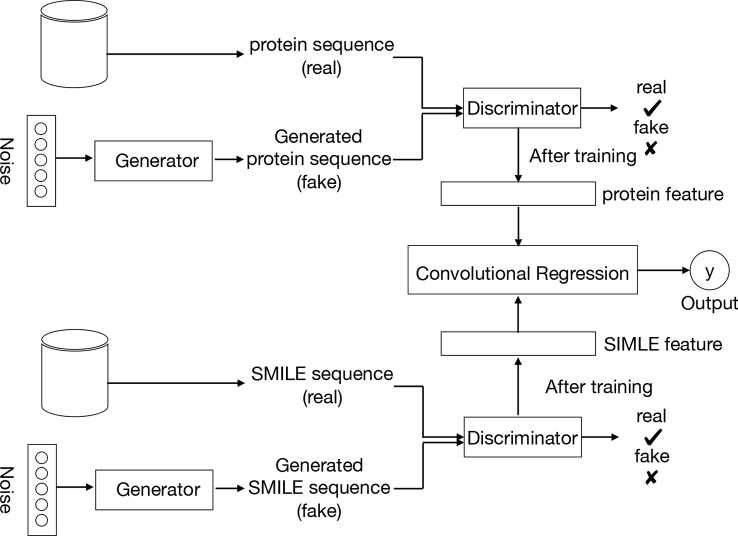
Figure 2.
Pipeline overview. We train the GANs on the unlabeled data set. Compound SMILES and protein sequences are encoded and two independent GANs are applied to generate the fake samples. The trained discriminator of the GANs can then be used to project the labeled data sets into a feature latent space. Based on this feature, we train a convolutional regression to predict the DT binding affinity.