Figure 1.
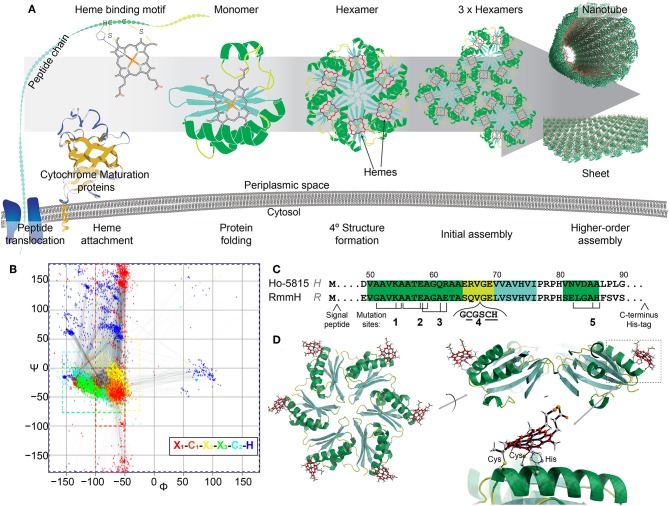
Design of heme-functionalized BMC-H shell proteins. (A) Cartoon schematic showing a hypothetical pathway of a heme from attachment on a shell protein to the formation of large structures. (B) Ramachandran plot of torsion angles within the heme binding motifs of naturally occurring cytochrome c structures (n = 249 heme binding motifs). Each color represents one amino acid in the motif. (C) Partial protein sequences of Ho-5815 and RmmH shell proteins. Secondary structures are highlighted (green: helix, yellow: loop, cyan: sheet). Three amino acid residues (black ticks) at Mutation sites 1,2,3, and 5 were changed to generate a heme binding motif (CxxCH). Six amino acids were inserted at mutation site 4 to introduce the heme binding motif at the loop region. (D) Molecular dynamics model of BMC-H variant R1 (heme at Mutation site 1 in RmmH). Conjugated carbon atoms in hemes are shown in red.