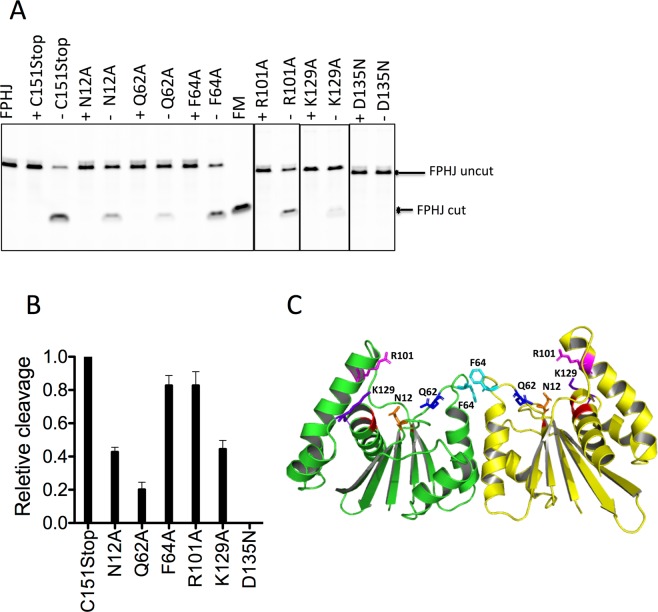
Figure 2.
HJ resolution activity of select Fpr C151Stop mutants. (A) Cleavage of fluorescently labeled FprHJ DNA by Fpr with amino acid substitutions for residues near the active site, monitored on denaturing gels. The reaction mixture (20 μl) containing Fpr (300 nM) with FprHJ (100 nM) was incubated at 37 °C for 10 min. FprHJ, FprHJ only in reaction buffer as a control. FM, FprHJ marker (nicked duplex product). For each protein a sample containing 5 mM EDTA (+) served as the negative control, and that containing 5 mM MgCl2 without EDTA (−) shows the enzymatic activity. Borders of cropped gels are highlighted by black lines. (B) Quantitation of the cleavage products of above mutants relative to that of Fpr C151Stop. (C) Structure of the Fpr dimer with side chains of the mutated residues shown. Backbone of the catalytic residues are colored in red. Supplementary Figure S2 shows activities of the other mutants tested. Each DNA cleavage experiment was repeated 3 times and a representative cropped gel is shown in the figure. Original full-length gels for Fig. 2 are shown in Supplementary Figure S10.