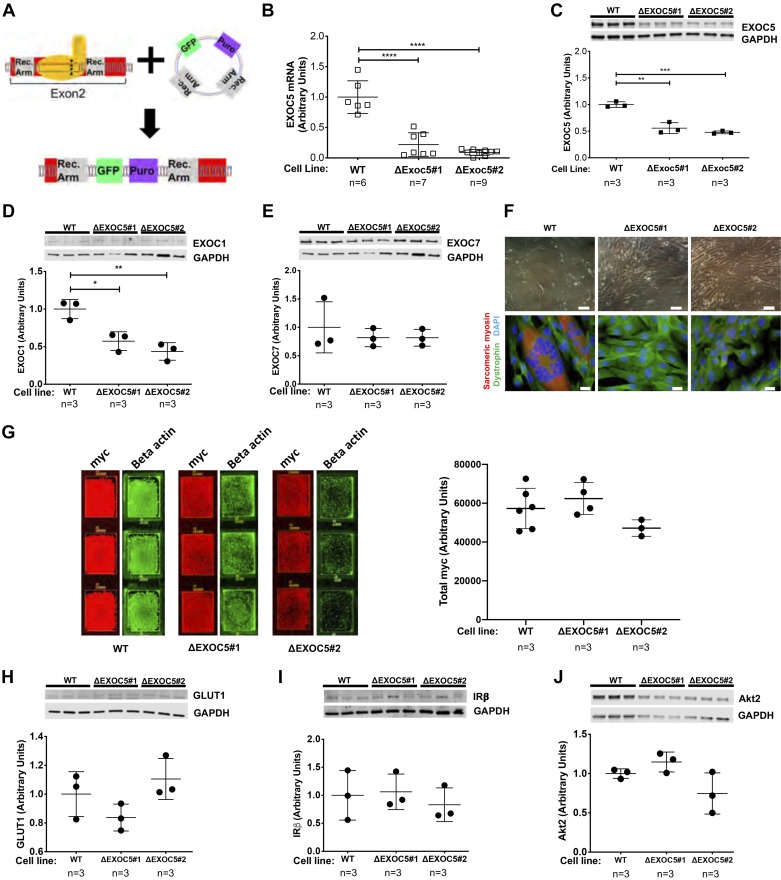
Fig. 6.
Heterozygous knockout of EXOC5 decreases EXOC1 subunit expression, impairs myotube differentiation, but does not affect expression of insulin signaling components. A: our approach to gene editing using CRISPR/Cas9 mediated homologous recombination resulted in the insertion of a green fluorescent protein (GFP) reporter and a puromycin resistance cassette into exon 2 of the EXOC5 gene. B: quantification of EXOC5 mRNA levels in ΔExoc5 L6-GLUT4myc clonal myoblasts using quantitative real-time PCR. C–E: quantification of exocyst subunit EXOC1, EXOC5, and EXOC7 protein levels in ΔExoc5 and wild-type (WT) L6-GLUT4myc myoblasts using Western blot analysis. F: assessment of differentiation ability in ΔExoc5 and WT L6-GLUT4myc myoblasts using light microscopy and immunofluorescent staining. Myotube formation was indicated by the formation of multinucleated, sarcomeric myosin-positive (red) structures. Dystrophin (green) and DAPI (blue) were used to visualize the cell membrane and nucleus, respectively. Double scale bars = 100 µm; single scale bars = 10 µm. G: representative immunofluorescent staining and quantification of total GLUT4myc levels in permeabilized L6 GLUT4myc myoblasts. Signal intensities of entire surface of the chamberslide wells were measured with a fluorescent scanner, and total GLUT4myc levels were normalized to β-actin levels measured. H–J: quantification of GLUT1, insulin receptor β-subunit (IRβ), and Akt2 protein levels, respectively, in ΔExoc5 and WT L6-GLUT4myc myoblasts using Western blot analysis. Images and data are representative of 3 independent experiments. Values are means ± SD in relative units (vs. WT L6-GLUT4myc cells); n, no. of replicates. *P < 0.05. **P < 0.01. ***P < 0.005. ****P < 0.001.