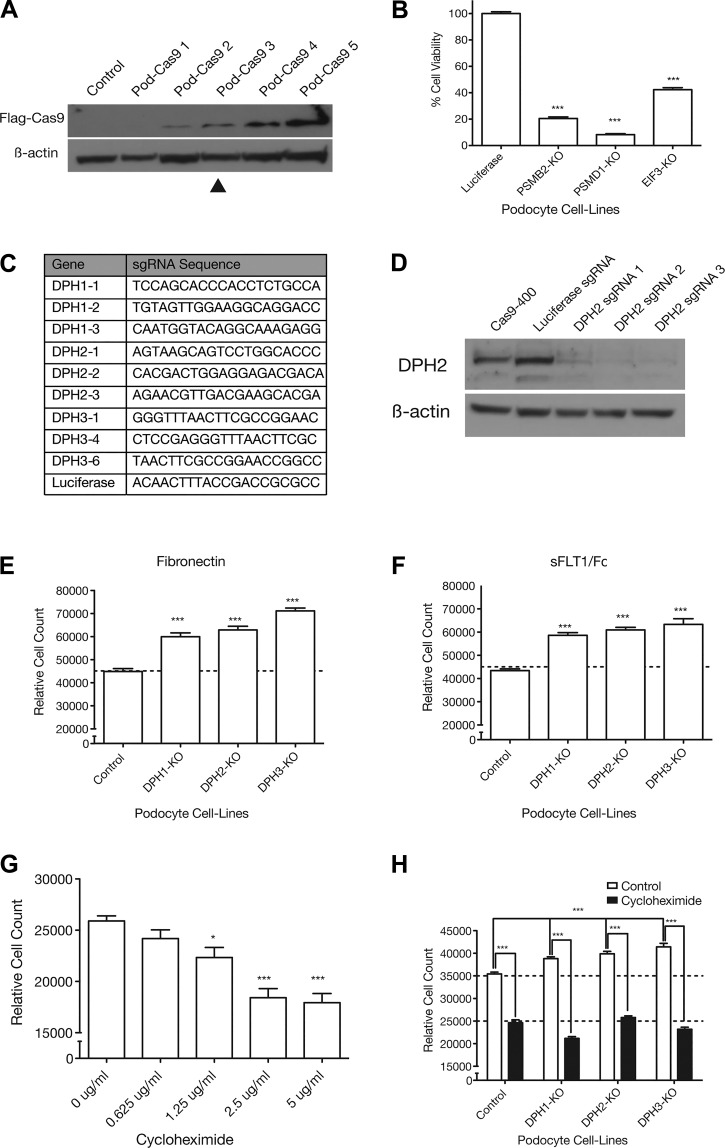
Fig. 3.
The increased adhesion phenotype in podocytes can be recapitulated using lentiCRISPR knockout (KO) of diphthamide biosynthesis genes and requires protein translation. A: Western blot showing increasing Cas9 expression levels in different Cas9-expressing podocyte cell lines. β-Actin was used as a loading control. B: previously validated single guide RNAs (sgRNAs) targeting essential genes [proteasome subunit-β2 (PSMB2), proteasome 26S subunit, non-ATPase 1 (PSMD1), and eukaryotic initiation factor 3 (EIF3)] were used to quantify editing efficiency in the Pod-Cas9-3 cell line by lethality. ***P < 0.001. C: guide sequences for three sgRNAs against each of diphthamide biosynthesis protein 1, 2, and 3 (DPH1, DPH2, and DPH3) and a luciferase control. D: DPH2 editing efficiency was tested in Pod-Cas9-3 cells by infecting three different sgRNAs against DPH2 at low multiplicity of infection. Western blot for DPH2 showed almost complete knockout of DPH2 protein with all three sgRNAs. E and F: three sgRNAs targeting each of DPH1, DPH2, and DPH3 were expressed in Pod-Cas9-3 cells to validate the adhesive phenotypes observed in the screen. Separate guide data were consolidated into one data point for easier visualization. (Individual guide data are available in Supplemental Fig. S1, A and B.) All DPH KO cell lines showed a statistically significant increase in adhesion to fibronectin and soluble Fms-like tyrosine kinase-1 (sFLT1)/Fc compared with control cells. ***P < 0.001. G: blockade of translation elongation by incubating podocytes overnight in cycloheximide caused a dose-dependent decrease in adhesion to fibronectin. H: blockade of translation by overnight incubation with 5 μg/mL cycloheximide in DPH1, DPH2, and DPH3 KO podocytes abolished the increased adhesive phenotype. Deconvoluted sgRNA data are provided in Supplemental Fig. S1F. ***P < 0.001; *P < 0.05.