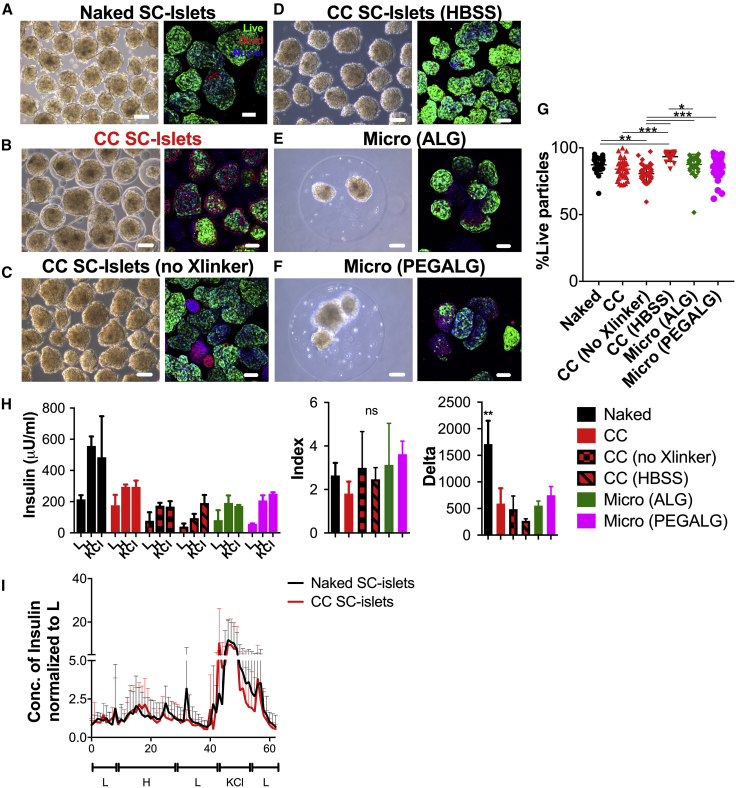
Figure 3.
Encapsulation Methods Comparison In Vitro
(A–G) Phase-contrast images (left) and live/dead confocal images (maximal projection of 150-μm-thick z stacks) of live (green)/dead (red) stained cells (nuclei: blue) (right) and quantification (individual data points represent the percent viability for each z level imaged) (G) of unencapsulated (A, naked), CC (using PEG and SH-PEG-SH Xlinker as pre-gel solution to resuspend SC islets before running them through the CC process) (B), CC no Xlinker (using PEG without SH-PEG-SH as solution unable to form gels to resuspend SC islets before running them through the CC process) (C), CC HBSS (using HBSS as solution to resuspend SC islets before running them through the CC process) (D), 1.2% MVG alginate (ALG) microencapsulated using electrostatic droplet generators (E), and 1.2% MVG alginate and PEG (PEGALG) (F) microencapsulated SC islets. Scale bars, 200 μm.
(H) Static GSIS functionality of naked (black), CC (red), CC no Xlinker (red with black squares), CC HBSS (red with black stripes), ALG microencapsulated (green), and PEGALG microencapsulated (fuchsia) S6d9 SC islets as absolute insulin secretion, index, and delta during sequential stimulation with 2.8 mM glucose (L), 20 mM glucose (H), and 30 mM KCl solutions. ∗∗p < 0.01 (n = 3 wells assayed per condition).
(I) Perifusion (100 μL/min perfusion with L: 2.7 mM glucose; with H: 20 mM glucose; with KCl: 2.7 mM glucose and 30 mM KCl solutions) of naked (black) and CC (red) SC islets normalized to L values. Four batches of naked and CC SC islets per condition. ns, no significant differences found.
All error bars are derived from standard deviations.