Figure 1.
Derivation of PPs from hPSCs Using Multiple Differentiation Protocols
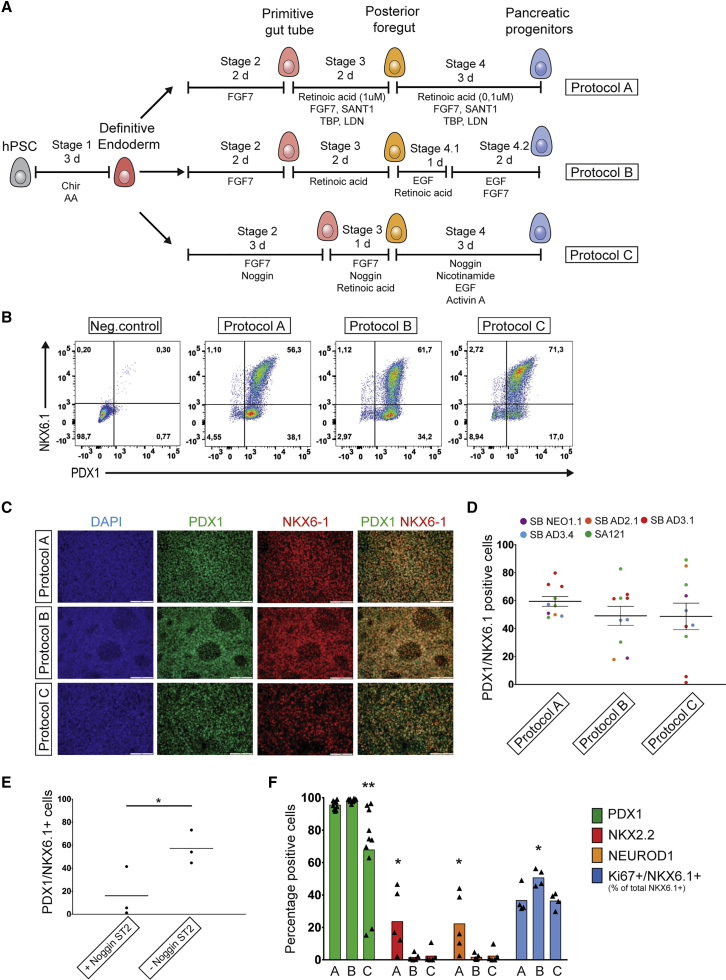
(A) Schematic outline of the three PP protocols applied.
(B) Representative examples of flow cytometry pseudo color dot plots of PPs from the three protocols stained for PDX1 and NKX6.1. Negative control is definitive endoderm cells.
(C) Immunofluorescence images of PPs stained for PDX1 and NKX6.1. Scale bar, 100 μm.
(D) Quantification of PDX1 and NKX6.1 co-expressing cells based on the flow cytometric analysis shown in (B). Graph shows a scatterplot of the mean ± SEM of five individual hPSC lines. Dots are color coded according to individual cell lines (details in Figure S1C). n = 10 independent experiments.
(E) Percentage of PDX1 and NKX6.1 co-expressing cells from the SB AD3.1 hiPSC line differentiated with protocol C with or without 50 ng/mL Noggin included during stage two; n = 3 independent experiments; ∗p < 0.05, paired t test.
(F) Quantification of PDX1, NKX2.2, NEUROD1, and percentage NKX6.1+ cells co-expressing Ki67. Bars show means and dots represent individual differentiations. One-way ANOVA with the Tukey test for multiple comparisons, ∗p < 0.05, ∗∗p < 0.01, different from the two other groups. PDX1, n = 10 independent experiments, same hPSC lines as in (D). NKX2.2 and NEUROD1, n = 5 independent experiments, one for each of the following hPSC lines: SA121 hESC, SB NEO1.1 hiPSC, SB AD2.1 hiPSC, SB AD3.1 hiPSC, SB AD3.4 hiPSC. Ki67, n = 4 independent experiments, three using SB AD3.1 hiPSC and one using SA121 hESC.