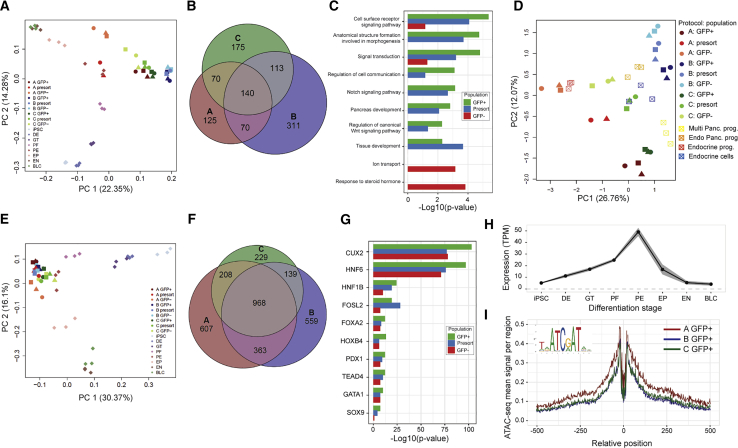
Figure 3.
Common Transcriptomic and Epigenomic PP Signatures across Three Differentiation Protocols
(A) PCA of RNA-seq samples together with data collected at all stages of the hPSC differentiation toward beta-like cells generated with protocol A (Perez-Alcantara et al., 2018).
(B) Venn diagram of the RNA-seq GFP+ PP signatures generated separately for each of the protocols by comparison with cells at the other differentiation stages.
(C) Selected gene ontology enrichment of the common PP signature genes across the three protocols; shown separately for signatures derived with presort cells (in blue), GFP+ (in green), and GFP− (in red) cell populations. The length of the bar represents −log10 of the enrichment p value.
(D) PCA of RNA-seq samples from this study, together with transcriptomes of fetal pancreas cell subpopulations (Ramond et al., 2018).
(E) PCA of ATAC-seq samples from this study, together with data collected at all stages of the hPSC differentiation toward beta-like cells, generated with protocol A (Perez-Alcantara et al., 2018).
(F) Venn diagram of the ATAC-seq GFP+ PP signatures generated separately for each of the protocols by comparison with cells at the other differentiation stages.
(G) Enrichment of selected TFs within the common PP signature open chromatin peaks across the three protocols; shown separately for signatures derived with presort cells (in blue), GFP+ (in green), and GFP− (in red) cell populations.
(H) Mean expression of CUX2 gene across all stages of hPSC differentiation toward beta-like cells. Shaded gray area indicates ± SEM (Perez-Alcantara et al., 2018).
(I) Footprinting analysis of CUX2 binding motifs within open chromatin peaks of the GFP+ populations generated with the three differentiation protocols.
BLC, beta-like cells; EN, endocrine cells; ; EP, endocrine progenitors; GT, gut tube; PE, pancreatic endoderm/progenitors; PF, posterior foregut.