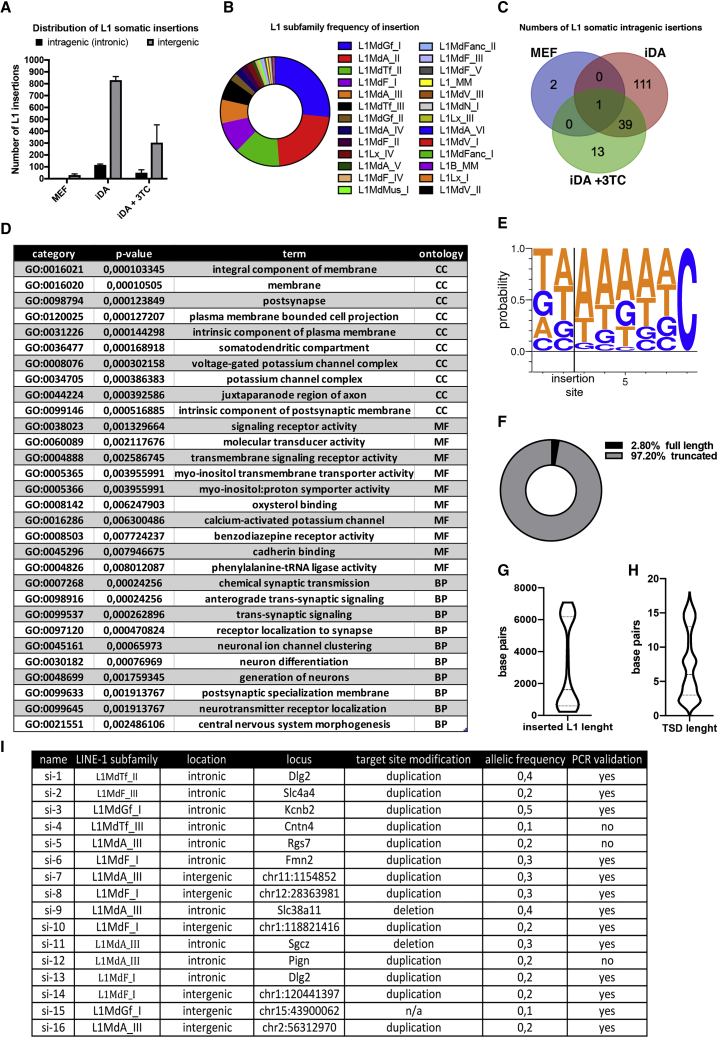
Figure 4.
Analysis of L1 De Novo Insertions in Reprogrammed and Lamivudine-Treated Cells
(A) Whole genome sequencing data of the distribution of L1 somatic insertions supported by more than five reads on each of 5’ and 3’ ends of inserted sites that were consistently inserted in more than two replicates between 0 and 50 base pairs in different replicates. SEM is indicated in the plot. n = 3.
(B) Representation of the frequency of retrotransposition of L1 subfamilies in iDA cells genome.
(C) Venn diagram showing different intragenic L1 de novo insertion rates.
(D) Functional analysis of L1 acceptor genes specific to MEF, iDA and 3TC-treated iDA cells using Gene Ontology.
(E) L1 insertion site motif LogoPlot.
(F) percentage of full length de novo inserted L1s.
(G) Truncation pattern of L1 somatic insertions.
(H) Target Size Duplication (TSD) size distribution. Table showing PCR validation results of 16 L1s somatic insertions.
(I) Selected L1 insertions used for PCR validation.
See also Figures S3 and S4.