Figure 5.
De Novo Insertions Support the Establishment of a Cell-Type-Specific Gene Expression Profile that Is Disrupted upon Lamivudine Treatment
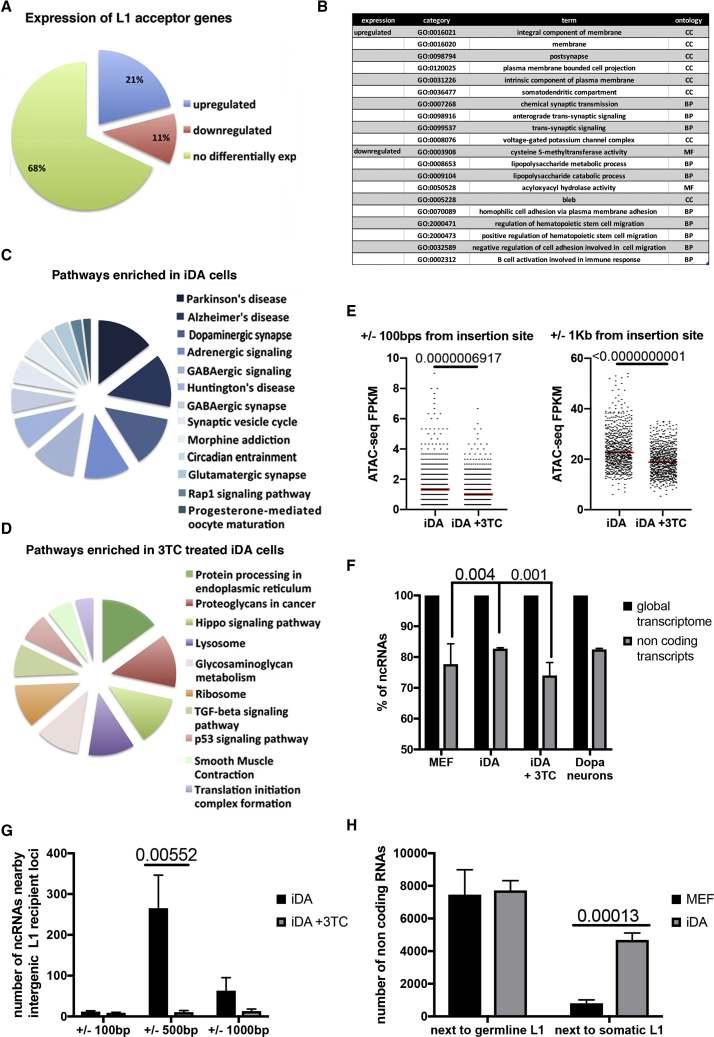
(A) Differential expression analysis of iDA-specific acceptor genes showing up, down, and not differentially expressed. False discovery rate (FDR) of 0.05 and a minimum fold change of 2 on a log2 scale have been considered in the analysis.
(B) Gene Ontology analysis of up- and down-regulated L1 acceptor genes.
(C) RNA-seq pathway analysis of differentially expressed pathways in iDA neurons compared with MEF cells.
(D) RNA-seq pathway analysis of differentially expressed pathways in 3TC-treated iDA neurons compared with mock iDA neurons.
(E) ATAC-seq analysis results at intergenic L1 recipient loci in iDA and 3TC treated cells at different distances from the insertion site; t tests are indicated. n = 3.
(F) Quantification of novel non-coding transcripts produced within ±100, ±500, ±1,000 bp from the somatic L1 element in iDA and 3TC-treated iDA neurons. SEM and t test are indicated. n = 3.
(G) Total amount of long non-coding transcripts produced in MEF and transdifferentiated cells within ±1,000 bp of de novo inserted and germline L1 elements. SEM and t test are indicated in the plot. n = 3.
(H) RNA-seq de novo assembly transcriptome analysis of MEF, iDA, dopaminergic neurons, and 3TC treated iDA cells. The percentage of non-coding transcripts longer than 200 bp has been normalized over the whole transcriptome. SEM and two-tailed t test are indicated in the plot. n = 6.
See also Figure S5.