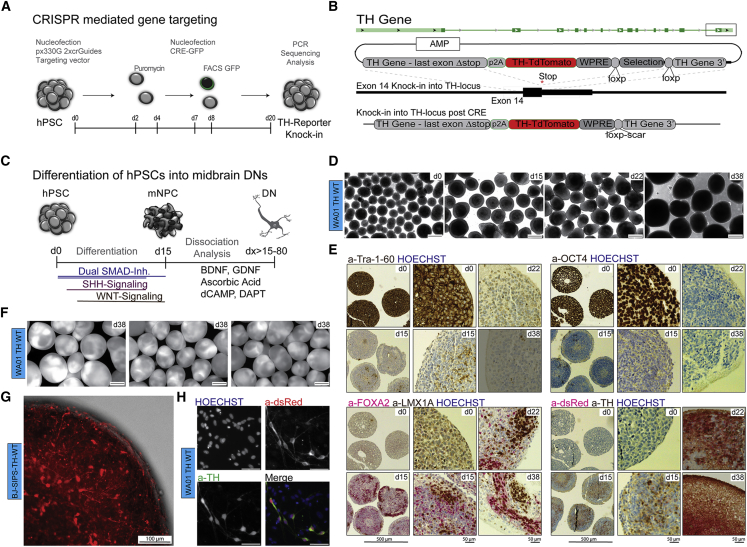
Figure 1.
Spin Culture Differentiation of TH Reporter hPSCs into Midbrain DNs
(A) Experimental scheme depicting the CRISPR-mediated TH reporter knockin strategy.
(B) Donor plasmid containing the targeting vector TH with a 5′TH homology arm followed by a 2A self-cleaving peptide sequence, a WPRE sequence, floxed selection cassette, and 3′ TH homology arm. Genomic locus indicating the targeting area in exon 14 of the TH gene, red star representing the position of the stop codon. Schematic representation of successful targeting post-CRE excision.
(C) Experimental scheme showing the culture conditions in spin culture.
(D) Bright-field images of WA01-WT spheres at time points d0, d15, d22, and d38. Scale bars, 500 μm (4×).
(E) Immunohistochemistry of sectioned organoids/spheres. Each panel organized as (top left, 4× d0; bottom left, 4× d15. Scale bar, 500 μm (4×); top middle, 20× d0; bottom middle, 20× d15; top right, 20× d22; and bottom right, 20× d38. Scale bar, 50 μm (20×) using validated antibodies against: top left panel (TRA-1-60), top right panel (OCT4), bottom left panel (FOXA2 and LMX1A), and bottom right panel (dsRed and TH). Nuclei were counterstained using HOECHST.
(F) Fluorescence images of WA01-WT spheres at d38 showing TH:TdTomato reporter expression. Scale bars, 500 μm, 4× (n = 3 differentiation experiments).
(G) Maximum projection image of d22 sphere from differentiated BJ-SIPS WT TH cells showing TH:TdTomato expression. z stack images acquired using spinning disc confocal CX7 at 10×. Scale bar, 100 μm.
(H) Fluorescence microscopy panel, showing DNs at d38 from WA01-TH-WT cell lines, dissociated on d25 and plated in low density on glial cells. Scale bars, 50 μm, 20× (top left, Hoechst; top right, a-dsRed; bottom left, a-TH [MAB318]; bottom right, merge).