Figure 3.
OS, Mitochondrial Dysfunction and Lysosomal Dysregulation Are Shared Phenotypes in all Early-Onset PD DNs
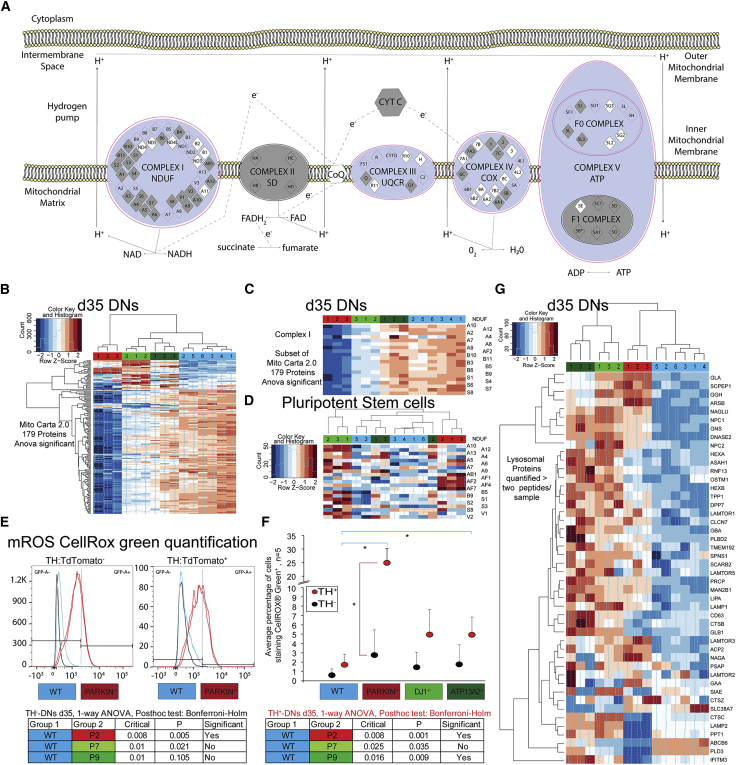
(A) The canonical oxidative phosphorylation pathway created using IPA software. log2 fold changes were plotted. Purple outlines indicate significantly dysregulated proteins/pathways (based on −0.7/0.7 log2 fold change). Red color indicates a positive change or protein enrichment in the PARKIN−/− line (none noted), while blue represents a depletion.
(B, D, G) Heatmap.2 function across all samples using specified gene lists, default clustering and row scaling. Columns represent samples, rows represent genes, and color intensity represents column Z score, where red indicates enriched and blue depleted proteins.
(B) Heatmap analysis of 179 Mito Carta 2.0 proteins with mitochondrial function or localization that shows differential protein abundance between any cell line in d35 DNs.
(C) Subset of Mito Carta 2.0 analysis showing all quantified proteins that are part of the NDUF-complex I in d35 DNs.
(D) Heatmap of all proteins that were quantified with at least two unique peptides that are part of the NDUF-complex I in hPSCs.
(E) Dissociated spheres at d35 were stained with CellROX green to detect mROS production in the basal state. mROS-G+ was quantified in TH− as well as TH+ cells. Histograms of mROS-G positivity overlaid with no staining control, stained WT (blue) and PARKIN−/− (red) lines, respectively.
(F) Averaged mROS percentage, quantified as mROS-G+ using synchronized bisector gates (FloJo, LLC) to divide the x axis into GFP− and GFP+ non-overlapping populations based on the background expression in our unstained control. Statistical significance was analyzed by one-way ANOVA followed by Bonferroni-Holm multiple comparison test (n = 4 independent staining and flow experiments significance indicated in tables [bottom]).
(G) Heatmap analysis of proteins with an annotated lysosomal function that were quantified with at least two unique peptides.