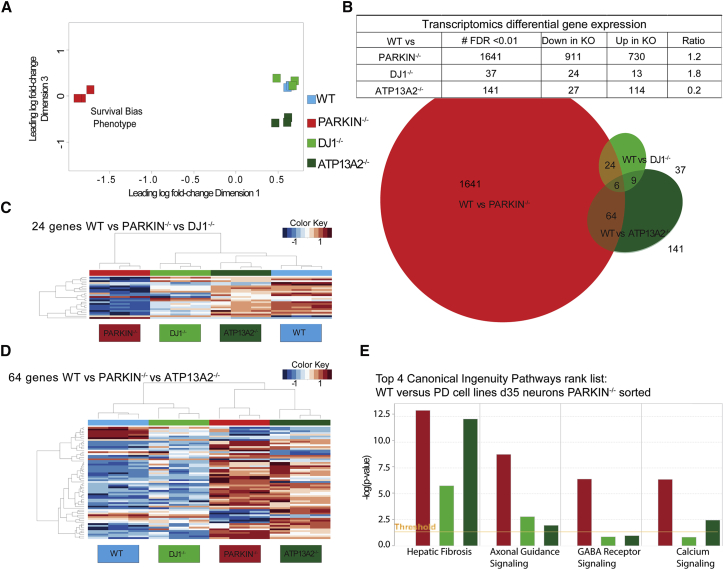
Figure 5.
Transcriptomics Analysis of Sorted DNs Highlights Common and Distinct Dysregulation of PD-Relevant Genes and Pathways: HUES1 PD Lines
(A) Multidimensional scaling plot of all isogenic cell lines in which distance corresponds to leading log fold changes between each pair of RNA samples. Dimension 1 separates PARKIN−/− line from all other lines and dimension 3 separates the ATP13A2−/− line from WT and DJ-1−/− lines.
(B) Table showing the number of significant DEGs between WT and isogenic lines (FDR = 0.01), the number of up- and downregulated genes, and the resulting ratio. Overlap analysis as a three-way Venn diagram intersecting DEGs between WT and all isogenic lines.
(C) Heatmap across all samples using the 24 intersecting genes between WT versus PARKIN−/− and WT versus DJ-1−/− analyses.
(D) Heatmap across all samples using the 64 intersecting genes between WT versus PARKIN−/− and WT versus ATP13A2−/− analyses.
(E) Bar chart of d35 global transcriptomics comparisons between WT and all PD cell lines showing select altered disease or biological function pathways determined using IPA curated sets. The y axis shows the significance (–log(p value)) and the orange line shows the significance threshold cutoff of −log(p value = 0.05), sorted by significance in PARKIN−/− versus WT comparison.