Abstract
Recently, a blaNDM-9 and mcr-1 co-harboring E. coli ST 617 isolate was identified from an asymptomatic carrier in Korea. An 81-year-old female was admitted to a university hospital for aortic cardiac valve repair surgery. Following surgery, she was admitted to the intensive care unit (ICU) for three days, and carbapenem-resistant E. coli YMC/2017/02/MS631 was isolated from a surveillance culture (rectal swab). Antimicrobial susceptibility testing (AST) for colistin was not performed at that time. Upon retrospective study, further AST revealed resistance to all tested antibiotics, including meropenem, imipenem, ceftazidime-avibactam, amikacin, gentamicin, ciprofloxacin, trimethoprim-sulfamethoxazole, and colistin, with the exception of tigecycline. Whole genome sequencing analyses showed that this strain belonged to the ST617 serotype O89/162: H10 and harbored three β-lactamase genes (blaTEM-1B, blaCTX-M-55, blaNDM-9), mcr-1, and 14 other resistance genes. Seven plasmid replicon types (IncB, IncFII, IncI2, IncN, IncY, IncR, IncX1) were identified. Horizontal transfer of blaNDM-9 and mcr-1 from donor cells to the recipient E. coli J53 has been observed. blaNDM-9 and mcr-1 were carried by IncB and IncI2 plasmids, respectively. To speculate on the incidence of this strain, routine rectal swab screening to identify asymptomatic carriers might be warranted, in addition to the screening of ICU patients.
Keywords: blaNDM-9, co-harboring Escherichia coli, mcr-1
1. Introduction
Since the first New Delhi metallo-β-lactamase 1 (blaNDM-1) was identified from a carbapenem-resistant Klebsiella pneumoniae strain [1], several variations of this metallo-β-lactamase have been discovered all over the world. One variant, blaNDM-9, was identified from a K. pneumoniae clinical isolate in China [2]. Accompanying the rise of carbapenem-resistant bacteria, colistin has become used more commonly, despite its serious side effects. Under the selective pressure of colistin, the first blaNDM-9 and mcr-1 co-harboring E. coli ST167 strain was identified from a retail chicken in 2016 [3]. Subsequent thereto, E. coli strains co-harboring blaNDM-9 and mcr-1 were reported from a chicken farm in China [4]. Since then, there have been concerns over the spread of this strain to humans. A blaNDM-9 and mcr-1 co-harboring E. coli 5CRE51 strain from a urine sample was first reported in Taiwan in 2017 [5]. This strain carried two different plasmids that harbored blaNDM-9 and mcr-1 and belonged to the ST617 strain [6]. Recently, a blaNDM-9 and mcr-1 co-harboring E. coli isolate was identified from an asymptomatic carrier in Korea in a retrospective study. Subsequent thereto, phenotypic and phylogenomic analyses were conducted to trace the strain’s origin.
2. Case Report
An 81-year-old female was admitted to a university hospital in February 2017 for aortic cardiac valve repair surgery. The patient had chest pain and dyspnea on exertion that had been aggravated six months before a diagnosis of severe aortic regurgitation. The patient had no history of abroad travel, visiting a farm or any signs of infections. She received preoperative antibiotic prophylaxis with amoxicillin-clavulanate. Following surgery, she was admitted to the intensive care unit (ICU) for three days, and carbapenem-resistant E. coli YMC/2017/02/MS631 was isolated from a rectal swab surveillance culture, which was routinely performed to stop the spreading of Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae in the ICU. Antimicrobial susceptibility testing (AST) for colistin was not performed at that time. Upon retrospective study, colistin resistance was detected: In vitro AST was performed using both broth microdilution and agar dilution methods. The results were interpreted according to the Clinical and Laboratory Standards Institute guidelines [7], with the exception of tigecycline and colistin, for which the European Committee on Antimicrobial Susceptibility Testing v9.0 was applied (http://www.eucast.org/clinical_breakpoints/). The isolate was resistant to all tested antibiotics (amoxicillin-clavulanic acid, piperacillin, piperacillin-tazobactam, cefotaxime, ceftazidime, cefepime, cefoxitin, aztreonam, ertapenem, meropenem, imipenem, ceftazidime-avibactam, amikacin, gentamicin, ciprofloxacin, trimethoprim-sulfamethoxazole, and colistin), except for tigecycline (Table 1).
Table 1.
Characterization of mcr-1, blaNDM-9, and blaCTX-M-55-positive E. coli YMC/2017/02/MS631 and its transconjugants.
| Parameters |
E. coli YMC/2017/ 02/MS631 |
Transconjugants | ||
|---|---|---|---|---|
| Selected by Colistin | Selected by Imipenem | E. coli J53 | ||
| E. coli EJ533 | E. coli EJ5331 | |||
| Source | Asymptomatic carrier | - | - | - |
| Isolation site | Rectal swab | - | - | - |
| Resistance genes |
blaNDM-9, blaCTX-M-55, blaTEM-1B, aph(3’)IIa, aph(3’)Ib, rmtB, aph(6)-Id, aadA2, oqxA, oqxB, fosA3, mph(A), mdf(A),floR, sul2, tet(A), dfrA12, mcr-1 |
mcr-1 |
blaNDM-9, aadA2, fosA3, mph(A), dfrA12 |
- |
| MLST | 617 | - | - | - |
| Serotype | O89/162:H10 | - | - | - |
| Plasmid replicon type(s) | IncB, IncFII, IncI2, IncN, IncY, IncR, IncX1 | IncI2 | IncB | - |
| Virulence factors | gad, iss | |||
| ompC, ompF | Intact | |||
| MIC (μg/mL, interpretation) | ||||
| Amoxicillin-clavulanic acid | 128, R † | 4, S ‡ | 8, R ‡ | 4, S ‡ |
| Piperacillin | ≥256, R † | N/D | N/D | N/D |
| Piperacillin-tazobactam | ≥256, R † | ≤4, S ‡ | ≥128, R‡ | ≤4, S ‡ |
| Cefotaxime | ≥256, R † | ≤1, S ‡ | ≥64, R ‡ | ≤1, S ‡ |
| Ceftazidime | ≥256, R † | ≤1, S ‡ | ≥64, R ‡ | ≤1, S ‡ |
| Cefepime | ≥256, R † | ≤1, S ‡ | ≥64, R ‡ | ≤1, S ‡ |
| Cefoxitin | ≥256, R † | 8, S ‡ | 32, R § | ≤1, S ‡ |
| Aztreonam | ≥128, R † | ≤1, S ‡ | ≤1, S ‡ | ≤1, S ‡ |
| Ertapenem | 64, R † | ≤0.5, S ‡ | 4, R ‡ | ≤0.5, S ‡ |
| Meropenem | 16, R † | N/D | N/D | N/D |
| Imipenem | 32, R † | ≤0.25, S ‡ | 8, R ‡ | ≤0.25, S ‡ |
| Ceftazidime-avibactam | ≥256, R † | N/D | N/D | N/D |
| Colistin | 4, R ‡ | 4, R ‡ | ≤0.125, S ‡ | <0.125, S ‡ |
| Amikacin | ≥16, R ‡ | ≤2, S ‡ | ≤2, S ‡ | ≤2, S ‡ |
| Gentamicin | ≥16, R ‡ | ≤1, S ‡ | ≤1, S ‡ | ≤1, S ‡ |
| Ciprofloxacin | ≥4, R ‡ | ≤0.25, S ‡ | ≤0.25, S ‡ | ≤0.25, S ‡ |
| Tigecycline | 0.5, S ‡ | ≤0.5, S ‡ | ≤0.5, S ‡ | ≤0.5, S ‡ |
| Trimethoprim-sulfamethoxazole | 320, R ‡ | ≤20, S ‡ | ≤20, S ‡ | ≤20, S ‡ |
Abbreviations: MIC, minimal inhibitory concentration; MLST, Multi Locus Sequence Type; N/D, not determined. In vitro antimicrobial susceptibility testing was performed using an agar dilution method † and a broth microdilution method ‡ following the Clinical and Laboratory Standards Institute (CLSI) guidelines M100 28th ed. MIC interpretations followed CLSI M100 28th ed, with the exception of tigecycline and colistin, for which the European Committee on Antimicrobial Susceptibility Testing guidelines v9.0 were applied.
Whole genomic DNA was extracted and sequenced using an Illumina Hiseq2500TM (Illumina, Valencia, CA, USA). Sequence reads were assembled using SPAdes version 3.12 [8]. The genome was annotated employing Rapid Annotation using Subsystem Technology [9] and deposited in the NCBI GenBank with the accession number SBHK00000000. Resistance genes, serotype, multi-locus sequence type (MLST), plasmid replicon type, and virulence factors were analyzed using ResFinder 3.1 [10], SeroTypeFinder 2.0 [11], MLST 2.0 [12], PlasmidFinder 2.0 [13], and VirulenceFinder 2.0 [14], respectively. Despite sharing the same sequence type (ST617) and serotype (O89/162: H10) with E. coli 5CRE51 [6], this strain carried additional resistance genes, namely, blaTEM-1B, blaCTX-M-55, aph(3’)IIa, aph(3’)Ib, rmtB, aph(6)-Id, oqxA, oqxB, mph(A), mdf(A), and sul2, accounting for cephalosporin, aminoglycoside, fluoroquinolone, macrolide, and sulfamethoxazole resistance (Table 1). Additionally, the mcr-1 sequence from E. coli YMC/2017/02/MS631 had one silent mutation at position 1074 (C→A), compared with a previously reported mcr-1. Seven plasmid replicon types (IncB, IncFII, IncI2, IncN, IncY, IncR, IncX1), and two virulence factors (gad, iss) were identified. Transferability of the blaNDM-9 and mcr-1 genes was achieved by conjugation experiments with E. coli J53 as the recipient strain [15,16]. Transconjugants were selected on Mueller–Hinton agar plates containing 100 μg/mL of sodium azide with 4 μg/mL of imipenem or 2 μg/mL of colistin. Resistance genes and plasmid replicon types in the transconjugants were identified by PCR and Sanger sequencing using designed primers (Table S1). The transconjugant E. coli EJ533 was susceptible to all tested antibiotics but colistin and harbored mcr-1 on the IncI2 type plasmid. Meanwhile, the transconjugant E. coli EJ5331 was resistant against all β-lactams but aztreonam and carried blaNDM-9 and other resistance genes (aadA2, fosA3, mph(A), sul2, and dfrA12) on the IncB type plasmid (Table 1).
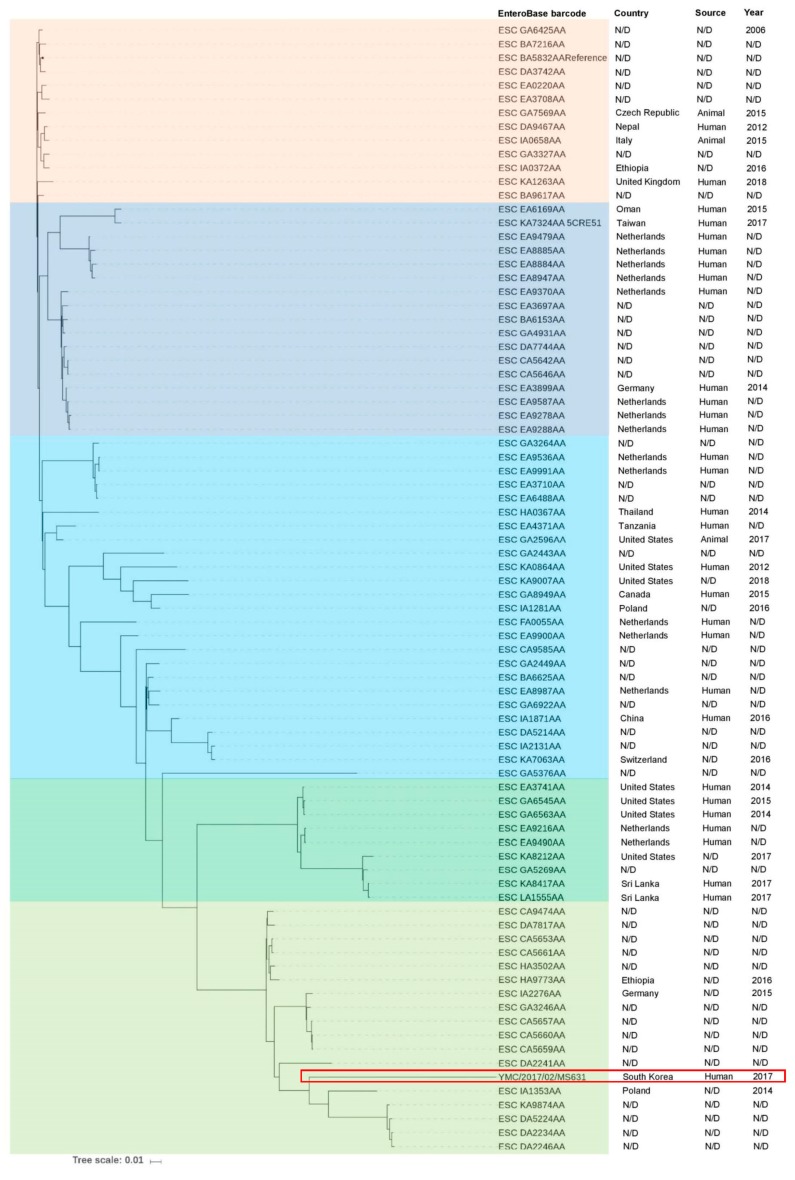
The relatedness of E. coli 5CRE51 and YMC/2017/02/MS631 in the context of worldwide E. coli ST617 distribution was investigated using EnteroBase database (http://enterobase.warnick.ac.uk/) (Figure 1). Altogether, 200 E. coli ST617 strains among 82,737 E. coli genomes (0.24%) have been reported in EnteroBase with the key words “E. coli” and “ST617” at the time of writing this paper (File S1). E. coli ST617 strains have been detected in at least 31 countries and from different sources, including human, animal, and environmental samples (Figure S1, S2A). Among them, 170 E. coli genomes were accessible for analysis (File S1). Thirteen different serotypes were identified from the collected genomes. Considering 170 genomes, 82 (48.2%) belonged to serotype O89/162: H10 (Figure S2B) and were included in the phylogenomic tree constructed using CSI phylogeny 1.4 [17]. The phylogenetic tree was visualized using iTOL (https://itol.embl.de/) and indicated that E. coli YMC/2017/02/MS631 was distant from E. coli 5CRE51 and other strains. The closest strains were detected in Poland (ESC_IA1353AA) and Germany (ESC_IA2276AA) (Figure 1). Meanwhile, E. coli 5CRE51 belonged to a clade with strains collected from Oman (ESC_EA6169AA) and The Netherlands (ESC_EA9479AA). This suggested that the two strains emerged independently.
Figure 1.
Phylogenomic comparison of the 82 E. coli ST617 strains collected from EnteroBase (http://enterobase.warnick.ac.uk/). The phylogenetic tree was constructed using CSI phylogeny 1.4 (https://cge.cbs.dtu.dk/services) with the standard parameters. iTOL (https://itol.embl.de/) was used to visualize the phylogenetic tree. The strain ESC_BA5832AA was selected as the reference strain. Origin, source and collection year of the isolates also were illustrated. The red box indicates the E. coli YMC/2017/02/MS631, which is closely related to the strain ESC_IA1353AA isolated from Poland in 2014.
3. Discussion
The first blaNDM-9 and mcr-1 co-harboring E. coli isolate belonged to a ST167 strain was followed by ST10 complexes, ST101, ST156, and ST297 [3,4]. The primary source was chicken meat. This strain rarely has been reported in clinical settings. Our report is important because it was detected coincidentally in a retrospective cohort study, and the strain displayed high antibiotic resistance with complicated plasmid types. Additionally, this strain displayed a broader resistance spectrum than the E. coli 5CRE51 isolate from Taiwan. Furthermore, the strain had two- and eight-folds higher minimal inhibitory concentrations (MICs) for meropenem and imipenem and an eight-fold higher MIC for amikacin in comparison with E. coli 5CRE51. However, the colistin MIC was the same: 4 μg/mL [5]. Resistome profiles, plasmid typing, and whole-genome phylogenetic tree analyses suggested that the two strains were not closely related. Interestingly, E. coli ST617 has mainly been detected in European countries, the United States, and China, and only two isolates of E. coli ST617 have been reported for Taiwan and Korea, according to EnteroBase (Figure S2A). The dominance of E. coli ST617 serotype O89/162:H10 among 13 serotypes may suggest that E. coli ST617 serotype O89/162: H10 can be a potential reservoir for blaNDM-9 and mcr-1 co-harboring E. coli in clinical settings.
According to the EUCIC medical guidelines on decolonization [18], there is a lack of evidence regarding the efficiency of decolonization for multidrug-resistant gram-negative organisms in hospitalized patients. Hence, the patient can be discharged without any treatment or intervention for decolonization. This may, in turn, lead to a silent continuous spread of this strain in the population at large. Additionally, travel by asymptomatic blaNDM-9 and mcr-1 carriers could facilitate strain transmission. A recent study emphasized the contribution of fecal pollution to an abundance of resistance genes in effluent-receiving environments [19]. Routine rectal swab screening to identify asymptomatic colistin and/or carbapenem-resistance carriers might be warranted to control the spread of multi-drug resistant organisms in hospital settings.
4. Conclusions
To the best of our knowledge, this is the first report of resistome profiles, plasmid typing, and whole-genome phylogenetic tree analyses of a blaNDM-9 and mcr-1 co-harboring E. coli strain isolated from an asymptomatic carrier. Moreover, the resistance genes were transferable to azide-resistant E. coli J53. Further surveillance studies should be conducted to detect the prevalence of this strain among health-care systems in Korea.
Abbreviations
AST: antimicrobial susceptibility testing; EUCIC: European Committee on Infection Control; ICU: Intensive care unit; MIC: Minimal inhibitory concentration; NCBI: National center for biotechnology information.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/8/4/212/s1.
Author Contributions
D.Y. and Y.L.C. designed the study and secured the funding. R.D., N.A.P., L.P.N., T.N.V., H.M., A.H.P. and H.L. performed the experiments. L.P.N., N.A.P., J.-H.B., R.D. and D.Y. analyzed and interpreted the data and wrote the manuscript.
Funding
This work was supported by the BioNano Health-Guard Research Center funded by the Ministry of Science, ICT & Future Planning (MSIP) of Korea as a Global Frontier Project (H-GUARD_2014M3A6B2060509); by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through Agricultural Microbiome R&D Program, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (918003-4); by a grant from the National Institute of Health, Korea (2019ER540300R514931). This work was also supported by the Brain Korea 21 PLUS Project for Medical Science, Yonsei University.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Data Availability
The genome YMC/2017/02/MS631 was deposited in the NCBI GenBank with the accession number SBHK00000000.
Ethics Approval and Consent to Participate
None.
References
- 1.Yong D., Toleman M.A., Giske C.G., Cho H.S., Sundman K., Lee K., Walsh T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009;53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X., Li H., Zhao C., Chen H., Liu J., Wang Z., Wang Q., Zhang Y., He W., Zhang F., et al. Novel NDM-9 metallo-β-lactamase identified from a ST107 Klebsiella pneumoniae strain isolated in China. Int. J. Antimicrob. Agents. 2014;44:90–91. doi: 10.1016/j.ijantimicag.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Yao X., Doi Y., Zeng L., Lv L., Liu J.-H. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect. Dis. 2016;16:288–289. doi: 10.1016/S1473-3099(16)00057-8. [DOI] [PubMed] [Google Scholar]
- 4.Liu B., Song F., Zou M., Zhang Q., Shan H. High Incidence of Escherichia coli Strains Coharboring mcr-1 and blaNDM from Chickens. Antimicrob. Agents Chemother. 2017;61:e02347-16. doi: 10.1128/AAC.02347-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai C.-C., Chuang Y.-C., Chen C.-C., Tang H.-J. Coexistence of MCR-1 and NDM-9 in a clinical carbapenem-resistant Escherichia coli isolate. Int. J. Antimicrob. Agents. 2017;49:517–518. doi: 10.1016/j.ijantimicag.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Lin Y.-C., Kuroda M., Suzuki S., Mu J.-J. Emergence of an Escherichia coli strain co-harbouring mcr-1 and blaNDM-9 from a urinary tract infection in Taiwan. J. Glob. Antimicrob. Resist. 2019;16:286–290. doi: 10.1016/j.jgar.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Wayne P. Performance Standards for Antimicrobial Susceptibility Testing Twenty-Eighth Informational Supplement M100-S28. 28th ed. Clinical and Laboratory Standards Institute; Wayne, NY, USA: 2018. [Google Scholar]
- 8.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M., et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joensen K.G., Tetzschner A.M.M., Iguchi A., Aarestrup F.M., Scheutz F. Rapid and Easy In Silico Serotyping of Escherichia coli Isolates by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2015;53:2410–2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larsen M.V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R.L., Jelsbak L., Sicheritz-Pontén T., Ussery D.W., Aarestrup F.M., et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carattoli A., Zankari E., García-Fernández A., Voldby Larsen M., Lund O., Villa L., Møller Aarestrup F., Hasman H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joensen K.G., Scheutz F., Lund O., Hasman H., Kaas R.S., Nielsen E.M., Aarestrup F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014;52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yi H., Cho Y.J., Yong D., Chun J. Genome sequence of Escherichia coli J53, a reference strain for genetic studies. J. Bacteriol. 2012;194:3742–3743. doi: 10.1128/JB.00641-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walsh T.R., Weeks J., Livermore D.M., Toleman M.A. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: An environmental point prevalence study. Lancet Infect. Dis. 2011;11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 17.Kaas R.S., Leekitcharoenphon P., Aarestrup F.M., Lund O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE. 2014;9:e104984. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tacconelli E., Mazzaferri F., de Smet A.M., Bragantini D., Eggimann P., Huttner B.D., Kuijper E.J., Lucet J.-C., Mutters N.T., Sanguinetti M., et al. ESCMID-EUCIC clinical guidelines on decolonization of multidrug-resistant Gram-negative bacteria carriers. Clin. Microbiol. Infect. 2019;25:807–817. doi: 10.1016/j.cmi.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Karkman A., Pärnänen K., Larsson D.G.J. Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nat. Commun. 2019;10:80. doi: 10.1038/s41467-018-07992-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome YMC/2017/02/MS631 was deposited in the NCBI GenBank with the accession number SBHK00000000.