Figure 2.
HDX-MS Analysis of HOIP/dAb Complexes
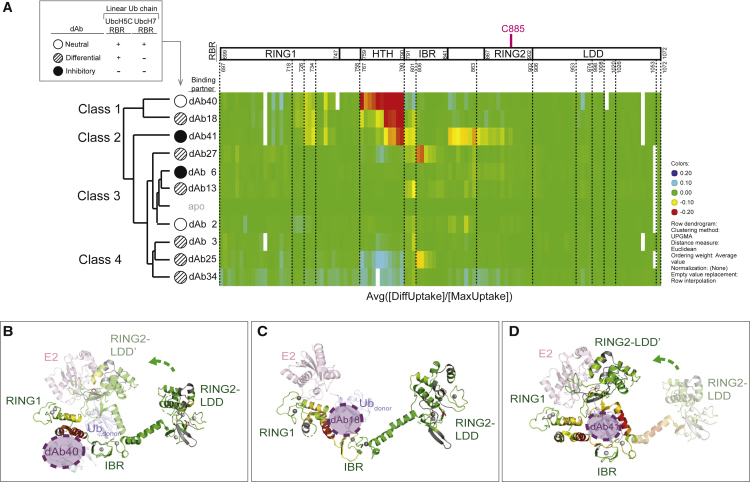
(A) Plot of differential deuteration versus HOIP peptides for each dAb complex. Four classes are presented according to clustering based on Euclidean distance approach. The functional effect of each dAb is symbolized as follows: white circle indicates neutral effect, circle with diagonal fill indicates differential effect, black circle indicates inhibitory effect.
(B–D) Differential deuteration heatmaps displayed on the structure of HOIP RBR in complex with a UbcH5B-Ub conjugate (PDB: 5EDV). The elongated HOIP RBR molecule found in the crystal structure is indicated by RING2-LDD, whereas the closed form that is suggested to bind UbcH5B-Ub is indicated by RING2-LDDʹ (Lechtenberg et al., 2016). The position of catalytic C885 located in RING2 is indicated in magenta stick representation. Structures are colored according to the deuteration heatmap shown in (A), gray indicates regions for which no peptides were recovered. (B) Example of a neutral dAb (dAb40) binding to a region of HOIP that has been suggested to bind an allosteric ubiquitin molecule (Lechtenberg et al., 2016). (C) Example of a differential dAb (dAb18) contacting a region close to the IBR domain that overlaps with the binding site of the donor ubiquitin. (D) Example of an inhibitory dAb (dAb41), which may trap the RBR domain in a conformation where the catalytic cysteine C885 is occluded.