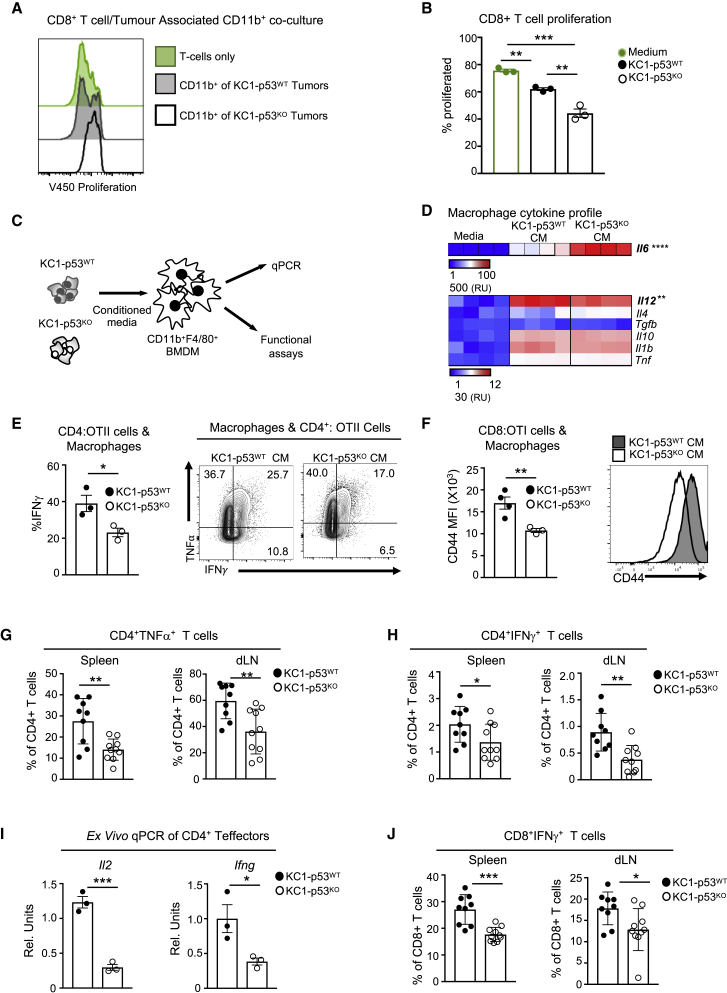
Figure 3.
Tumoral Loss of p53 Promotes Suppressive Myeloid Lineages and Reduces T Cell Activation
(A) CD11b+ cells isolated from individual KC1-p53WT (gray shade) and KC1-p53KO (black line) tumors 3 days post-injection and co-cultured with pre-activated CD8+ T cells stained with V450 proliferation dye. Cells were analyzed after 48 h. T cell proliferation in the absence of CD11b+ cells is denoted in green.
(B) Graph displays percent proliferation of CD8+ T cells with tumor-derived CD11b+ cells isolated from individual KC1-p53WT (black) or KC1-p53KO (white) tumors or in the absence of CD11b+ cells (green). Each dot represents CD11b+ cells derived from a pool of two tumors per genotype and means are represented as ±SEM.
(C) Schematic representation of the experimental design. BMDMs activated with conditioned media (CM) from KC1-p53WT or KC1-p53KO cells and screened for cytokine expression by qPCR and functional assays.
(D) mRNA expression of T-cell-polarizing cytokines expressed by BMDMs incubated in the presence of IMDM (medium), or KC1-p53WT, or KC1-p53KO conditioned media. Red indicates higher and blue lower expression levels, where each lane represents BMDMs derived from individual mice; the means are represented as ±SEM.
(E and F) Incubation of BMDMs with conditioned media, followed by a 2-h pulse of ovalbumin and co-cultured with CD4+OTII or CD8+OTI cells, respectively. Graphs show one of three experiments and display technical replicates (n = 3-4); means are represented as ±SD.
(E) OTII CD4+ T cells restimulated with OVA 323-339 peptide after co-culture with BMDMs educated by KC1-p53WT CM (closed circles) or KC1-p53KO CM (open circles). Representative flow cytometry plot (right) of restimulated OTII T cells followed by intracellular cytokine staining (ICS) for IFN-γ and TNF-α after 4 days of differentiation.
(F) CD8+OTI cells activated by BMDMs pulsed with OVA and measured by flow cytometry for surface expression of CD44, shown as mean fluorescence intensity (MFI). Left graph represents technical replicates for CD44 MFI, and on the right, the histogram shows CD44 surface expression.
(G–J) Analysis of CD4+ and CD8+ T cells in the periphery of FVB recipients injected with KC1-p53WT (black) or KC1-p53KO (white) cell lines. Graphs show biological replicates and the means are represented as ±SEM.
(G and H) Intracellular cytokine flow cytometry analysis of spleen and draining lymph nodes of FVB mice bearing KC1-p53WT and KC1-p53KO tumors (n = 9–10 FVB per genotype), 7 days post injection. Graphs illustrate (G) CD4+ T cells expressing TNF-α and (H) CD4+ T cells producing IFN-γ upon ex vivo restimulation with PMA, ionomycin, and GolgiStop (n = 9–10 per genotype).
(I) CD4+ T cells isolated from KC1-p53WT and KC1-p53KO tumor-bearing FVB mice and cell sorted for CD4+ CD25− populations. qPCR was performed on RNA isolated from ex vivo sorted CD4+ CD25− T cells and tested for Il2 and Ifng mRNA. The means are represented as ±SEM, and each point represents two pooled mice (cohort size n = 6 per genotype).
(J) Ex vivo restimulation of spleen and dLN from tumor bearing FVB mice to detect CD8+T cells producing IFN-γ. The means are represented as ±SEM (cohort size n = 9–10 per genotype).
Unpaired t tests were performed on all data except for multiple comparisons, where Tukeys’s multiple comparisons test was used. p values are ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. See also Figure S3.