Figure 3.
Comparison of Murine HGSOC and ICGC Ovarian Transcriptomes
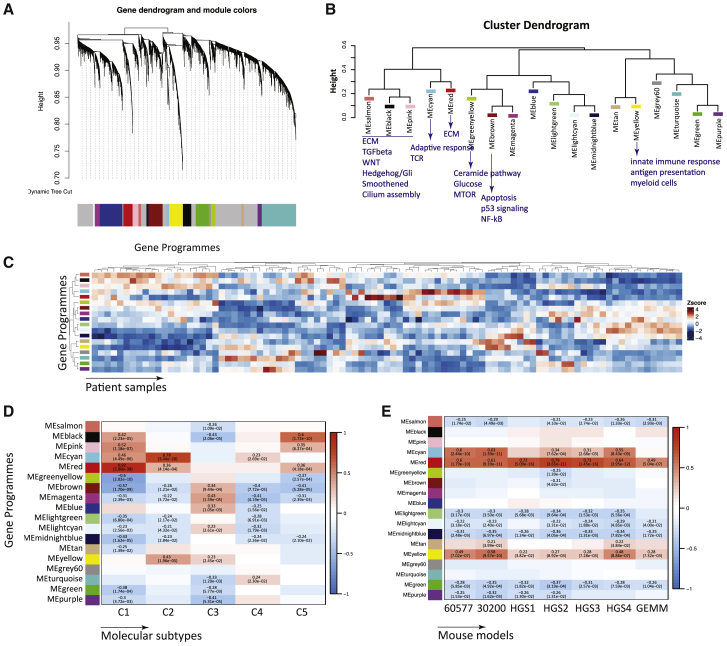
(A) Weighted correlation network analysis (WCNA) of human ICGC transcriptional HGSOC dataset showing clusters of co-regulated genes as a dendrogram. Colors show different modules (gene programs).
(B) Cluster dendrogram of module eigenvalues (MEs) illustrates clustering of programs associated with ECM, immune response, or tumor-related signaling pathways.
(C) Heatmap of MEs across ICGC samples (n = 93).
(D) Heatmap of association of C1–C5 classification. Positive associations are shown in red and negative associations are shown in blue. Pearson’s r and p values are indicated in the fields where a significant association was observed (p < 0.05).
(E) Heatmap of association of differentially expressed gene scores in mouse models. Positive associations are shown in red and negative associations are shown in blue. Pearson’s r and p values are indicated in the fields where a significant association was observed (p < 0.05).