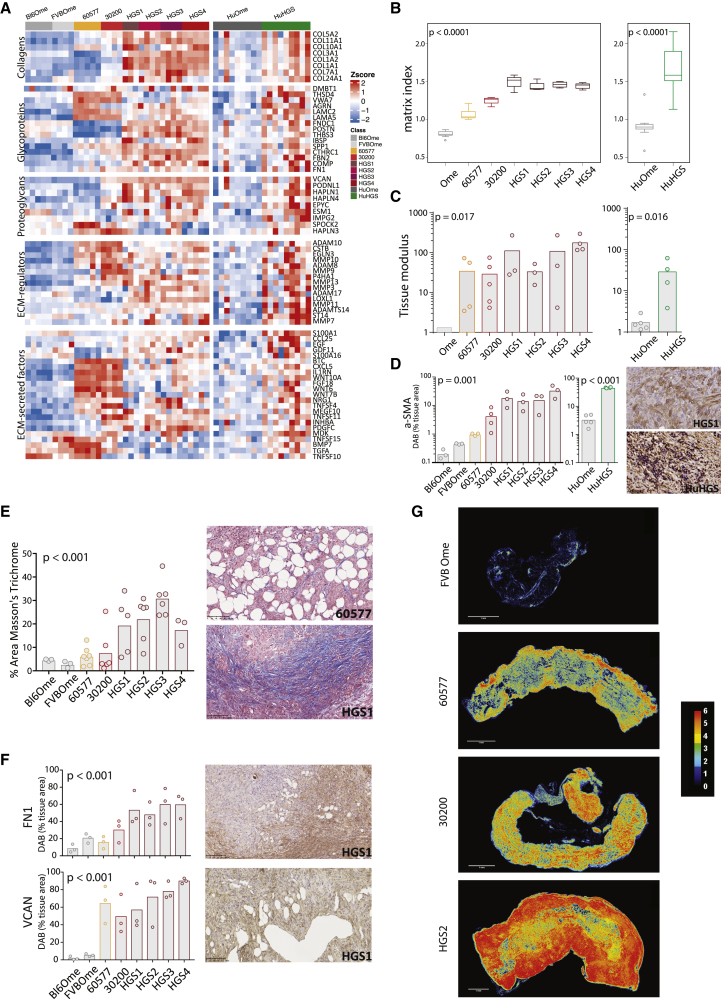
Figure 5.
Matrisome, Matrix Index, and Stiffness of Murine and Human HGSOC
(A) Heatmap of orthologous matrisome genes, grouped by matrisome class, in the mouse and human peritoneal datasets compared to normal omenta (Student’s t test, p < 0.05).
(B) Matrix Index of murine and human peritoneal HGSOC and normal omenta. 30200, HGS2 and HGS3, n = 4; 60577 and HGS4, n = 5; HGS1 n = 3; FVBOme n = 4; Bl6Ome n = 5. For human samples, n = 9 normal/adjacent omenta and n = 9 omental tumors.
(C) Tissue modulus of murine and human peritoneal HGSOC and normal omenta. Each dot represents a tumor from an individual mouse or patient. p values correspond to the Kruskal-Wallis test for mouse data and the Mann-Whitney U test for human data.
(D) Fibroblast content of murine and human peritoneal HGSOC and normal omenta was assessed by IHC for αSMA staining and quantified using the Definiens Tissue Studio platform. p values correspond to one-way ANOVA. Each dot represents a tumor from an individual mouse. A representative image for HGS1 is depicted at right. Scale bar, 50 μm.
(E) Masson’s trichrome staining was performed on all HGSOC model tumors and on normal omenta. Bar plot illustrates the result of digital analysis and the quantification of the percentage of positive area by the Definiens Tissue Studio. Representative images for 60577 and HGS1 are depicted at right. Scale bars, 100 μm. p values correspond to one-way ANOVA. Each dot represents a tumor from an individual mouse.
(F) IHC for FN1 and VCAN staining quantified using the Definiens Tissue Studio platform. p values correspond to one-way ANOVA. Each dot represents a tumor from an individual mouse. Representative images for HGS1 are depicted at right. Scale bars, 100 μm.
(G) Construction of tissue matrisome heatmaps for models of HGSOC. Serial IHC images were color deconvoluted, overlaid, and pseudo-colored using ImageJ to highlight areas that were rich (red) or poor (black) in ECM. Expression hotspots for all six ECM molecules are shown in red, whereas areas expressing one to five ECM molecules are presented with the different colors on the key map shown at right. Scale bars, 1 mm.