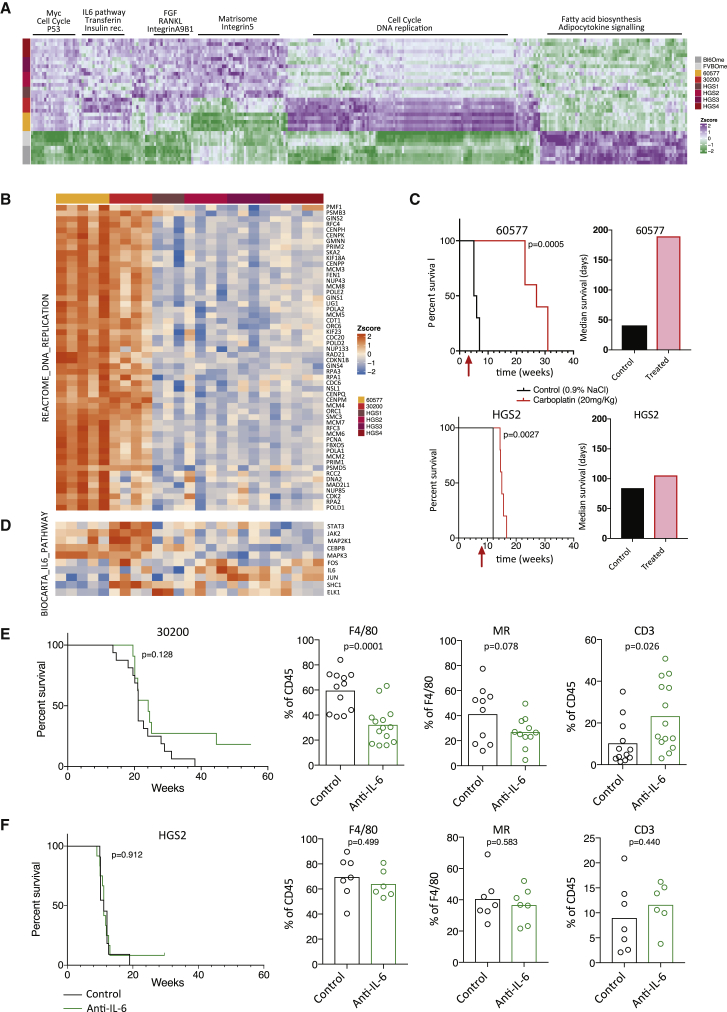
Figure 6.
Therapeutic Vulnerabilities of HGSOC Models
(A) Single-sample gene set enrichment analysis (GSEA) was performed for the murine HGSOC transcriptomes. Heatmap illustrates pathway scores with distinct expression patterns in 30200, 60577, HGS tumors, and normal omenta (FDR < 0.05). 30200, HGS2, and HGS3, n = 4; 60577 and HGS4, n = 5; HGS1, n = 3; FVBOme, n = 4; Bl6Ome, n = 5.
(B) Heatmap of REACTOME DNA replication pathway genes across the murine tumors.
(C) Response of mice injected with 60577 or HGS2 to three cycles of chemotherapy (carboplatin 20 mg/kg, once per week). Survival curve and median survival are shown (n = 5 mice per group). The log rank p value is depicted on the survival curves. The start of the treatment is indicated by the red arrow.
(D) Heatmap of BIOCARTA IL-6 pathway genes across the murine tumors.
(E and F) Mice injected with 30200 (E) or HGS2 (F) were treated with isotype control or anti-IL-6 i.p. 2 mg/kg twice weekly starting 10 (30200) or 7 (HGS2) weeks after cell injection until endpoint. The log rank p value is depicted on the survival curves (for 30200, ncontrol = 16 and ntreated = 11; for HGS2, ncontrol = 11 and ntreated = 12). Analysis of the immune infiltrate was performed by flow cytometry on a different set of mice and Student’s t test value is depicted on the bar plots. Each dot represents a tumor from an individual mouse. For 30200, two experiments pooled together are shown.