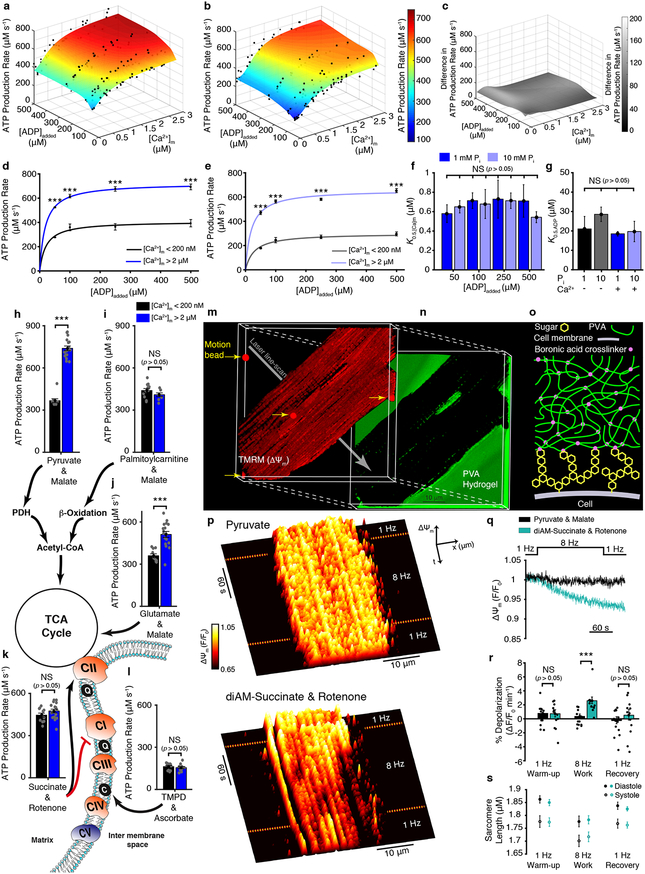
Figure 1. [Ca2+]m sensitive ATP production by mitochondria.
a. The dependence of ATP production (μM/s) on [Ca2+]m and [ADP]added at 1 mM Pi (n= 37, 38, 38, 29 for 500, 250, 100, 50 μM ADP added, respectively). b. Same as (a) but at 10 mM Pi (n= 28, 27, 30, 42 for 500, 250, 100, 50 μM ADP added, respectively). c. The difference in ATP production rates between 1 mM Pi and 10 mM Pi. d. Dependence of ATP production (μM/s) on ADP at 1 mM Pi when [Ca2+]m is < 200 nM (black circles, n= 3, 3, 3, 5 for 50, 100, 250, 500 μM ADP added, respectively) or > 2 μM (blue circles, n= 8, 7, 6, 9 for 50, 100, 250, 500 μM ADP added, respectively). Data are fit to a Michaelis–Menten equation. e. Same as (d) but at 10 mM Pi when [Ca2+]m is <200 nM (grey circles, n= 8, 6, 3, 7 for 50, 100, 250, 500 μM ADP added, respectively) or 2 μM (light blue circles, n= 5, 7, 5, 5 for 50, 100, 250, 500 μM ADP added, respectively). Data are fit to Michaelis–Menten equation. f. [Ca2+]m at which ATP production rate is half maximal (K0.5,[Ca]m) for [ADP]added at both 1 and 10 mM Pi. Each bar shows the K0.5,[Ca]m constant of each of the eight fit lines shown as surface plots in a-b (K0.5,[Ca]m ± s.e. of fit in μM, fitted sample size is given in a-b, individual data points shown in a-b). g. [ADP]added at which ATP production is half maximal (K0.5,ADP) for [Ca2+]m <100 nM (−) and >2 μM (+) at both 1 and 10 mM Pi. Each bar shows the K0.5,ADP constant of each of the four fit lines shown in d-e (K0.5,[Ca]m ± s.e. of fit in μM, fitted sample size is given in d-e, individual data points shown in a-b). h-l. ATP production rate at low [Ca2+]m (<200 nm, black bar) and high [Ca2+]m (>2 μM, blue bar) using the indicated combination of carbon substrates and metabolic inhibitors (n= 10–20 per group). Abbreviations used in the diagram: PDH, pyruvate dehydrogenase; TCA, tricarboxylic acid; CI, Complex 1; CII, Complex 2; CIII, Complex 3; CIV, Complex 4; CV, Complex 5 (i.e. ATP synthase); Q, ubiquinone; C, cytochrome c. m. 3D reconstruction of confocal Z-stack images of a cardiomyocyte loaded with the fluorescent indicator TMRM (tetramethylrhodamine methyl ester perchlorate, 50 nM). n. The fluorescence of fluorescein-containing poly(vinyl alcohol) (PVA) hydrogel that embeds the cell shown in (m). o. Diagram showing boronic acid crosslinker linking cell-surface sugars to the PVA hydrogel. p. Fluorescence surface plot demonstrating spatiotemporal changes of ΔΨm. Measurements are done on cardiomyocyte paced by field-stimulation to contract at 1 or 8 Hz in a bath (extracellular) solution that contains top; pyruvate (1 mM) and malate (0.5 mM), or bottom; diAM-succinate (succinic acid diacetoxymethyl ester, 10 μM) and rotenone (5 μM). q. Average ΔΨm fluorescence time course using pyruvate + malate (black, n = 13 cells) and diAM-succinate + rotenone (turquoise, n = 12 cells). r. Quantification of ΔΨm depolarization expressed as percent change per minute. s. Sarcomere length measured simultaneously in the experiments shown in q. The sample size (n) in all panels represents the number of independent experiments. Data in d-e, h-l, and r-s are mean ± s.e.m. One-way two-tailed ANOVA with Bonferroni correction in d-g, and two-sample two-tailed t-test in h-l and r. * P < 0.05, ** P<0.01, *** P<0.001.