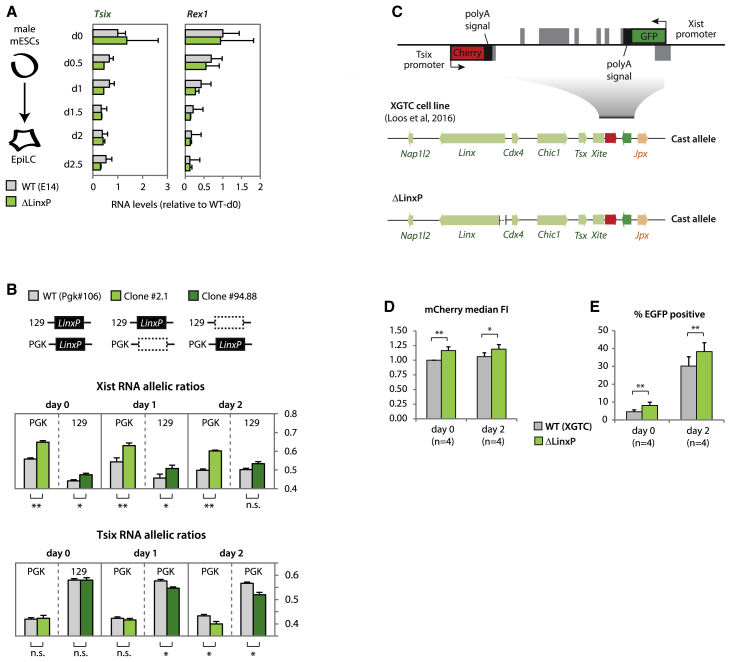
Figure 3.
The LinxP Element Is Not an Enhancer of Tsix but Regulates Xist Expression
(A) Gene expression analysis during differentiation. Data are normalized to wild-type day 0 for each gene, and represents the average of two biological replicates for each genotype.
(B) Allelic quantification of Xist (top) and Tsix (bottom) RNA during early differentiation. See legend of Figure 2C for more information on the clones. Data are presented as means and error bars represent SEM (six biological replicates). Statistical analysis was performed using a two-tailed paired t test with Bonferroni’s correction (∗∗p < 0.01).
(C) XGTC female line (129/Cast) as in Figure 1H. We generated ΔLinxP mutant clones on the Cast allele.
(D and E) Median fluorescence intensity (FI) of mCherry (normalized to WT, day 0) or percentage of EGFP positive cells (as in Figure 1J). Wild-type data represent an average of five wild-type clones, with four experimental replicates for each. ΔLinxP data represent an average of five independent clones, with four experimental replicates for each. Statistical analysis was performed using a paired two-tailed t test (∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001).