Figure 4.
Linx-Related Topological Features Are Not Implicated in Xist Regulation
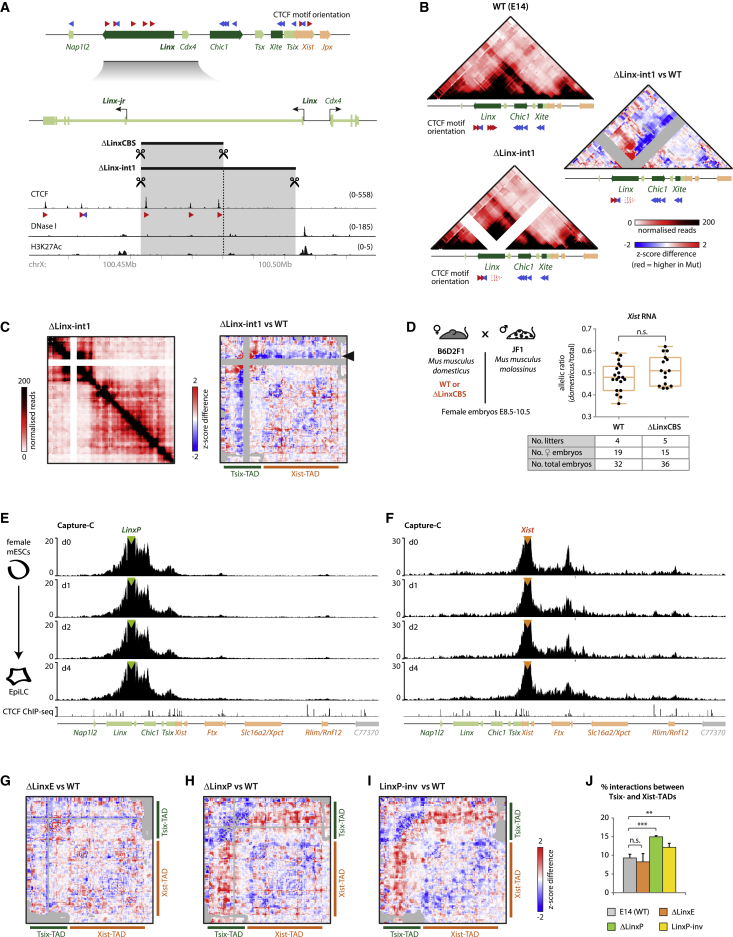
(A) The Linx locus, CTCF binding, and orientation of CTCF motifs associated with CTCF chromatin immunoprecipitation sequencing (ChIP-seq) peaks. Orientation of CTCF motifs within the Tsix-TAD is represented above. The targeted deletions ΔLinxCBS (∼25 kb) and ΔLinx-int1 (∼51 kb) are indicated. See STAR Methods for sources of CTCF, DNaseI, and H3K27Ac datasets.
(B and C) 5C profiles of the Tsix-TAD (B) and the two Xic TADs (C); pooled data from two biological replicates for each genotype. Differential map is corrected for deletion (see STAR Methods). Gray pixels represent either the deleted region or filtered contacts.
(D) Left: cross used for analysis of RNA allelic ratios in female hybrid embryos. Right: Xist RNA allelic ratios; each black dot corresponds to a single female embryo. Statistical analysis was performed using a two-tailed t test. The table summarizes the number of embryos collected. Analysis of Atp7a RNA allelic ratios and reverse cross is shown in Figures S5A and S5B.
(E and F) Capture-C profiles for LinxP (E) and Xist (F) viewpoints, at different time points of differentiation of XX (Pgk12.1) mESCs. Data represent one replicate; two or three replicates for each time point were performed and are identical to the one shown (data available in GEO). Profiles represent number of contacts for each DpnII fragment per 10,000 total contacts within a specified region (see STAR Methods). CTCF ChIP-seq on male mESCs is represented below (Nora et al., 2017).
(G–I) 5C differential maps for mutant male mESCs: ΔLinxE (G), ΔLinxP (H) and LinxP-inv (I); pooled data from two biological replicates for each genotype. 5C profiles for each genotype are shown in Figure S5D. Gray pixels correspond to either deleted regions or filtered contacts.
(J) Quantification of 5C inter-TAD contacts (see Figure S5E for details). Bars represent the average of the calculated proportions of four (E14 and ΔLinxP) or two (ΔLinxE and LinxP-inv) independent replicates. Statistical analysis was performed using a two-tailed t test (∗∗p < 0.01; ∗∗∗p < 0.001).