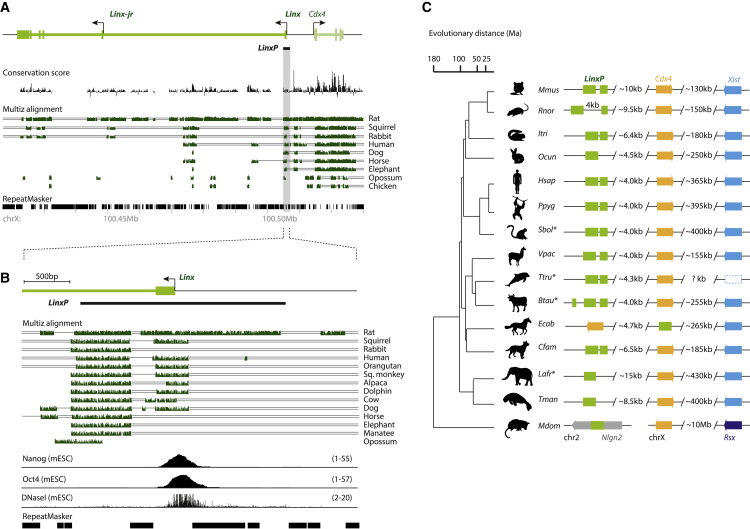
Figure 6.
The LinxP Element Is Conserved across Placental Mammals and Overlaps the Binding Site for Pluripotency Factors
(A) Sequence conservation analysis. Conservation score across placental mammals shows poor sequence conservation for Linx (compared to Cdx4), except for a few regions. Multiz alignment shows conserved stretches in green.
(B) Zoom-in from (A) of the Linx promoter region, showing two highly conserved modules across placental mammals. Nanog and Oct4 ChIP-seq, as well as DNaseI-seq (DNase I hypersensitive sites sequencing), are represented below (same as in Figure 2B)
(C) Synteny analysis across placental mammals and opossum of the two conserved modules identified in (B). Note that they are highly syntenic in placental mammals, lying close to Cdx4 and Xist on the X chromosome. In the marsupial opossum, the conserved element (half of one LinxP module) lies on chromosome 2, while Cdx4 and Rsx (the marsupial equivalent to Xist) lie on the X chromosome. Genomes of species marked with an asterisk (∗) are shown here in inverse orientation to what is annotated in UCSC for clarity purposes. Each species is designated by the first letter of its genus (in capital) and the first three letters of its specific epithet; the order of the species is the same as in (B), where they are designated by their common names. Evolutionary distance is represented in million years (Ma).